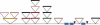A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion - PubMed (original) (raw)
Comment
A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion
K Shirasu et al. Genome Res. 2000 Jul.
Abstract
Organisms with large genomes contain vast amounts of repetitive DNA sequences, much of which is composed of retrotransposons. Amplification of retrotransposons has been postulated to be a major mechanism increasing genome size and leading to "genomic obesity." To gain insights into the relation between retrotransposons and genome expansion in a large genome, we have studied a 66-kb contiguous sequence at the Rar1 locus of barley in detail. Three genes were identified in the 66-kb contig, clustered within an interval of 18 kb. Inspection of sequences flanking the gene space unveiled four novel retroelements, designated Nikita, Sukkula, Sabrina, and BAGY-2 and several units of the known BARE-1 element. The retroelements identified are responsible for at least 15 integration events, predominantly arranged as multiple nested insertions. Strikingly, most of the retroelements exist as solo LTRs (Long Terminal Repeats), indicating that unequal crossing over and/or intrachromosomal recombination between LTRs is a common feature in barley. Our data suggest that intraelement recombination events deleted most of the original retrotransposon sequences, thereby providing a possible mechanism to counteract retroelement-driven genome expansion.
Figures
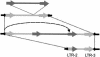
Figure 1
Arrangements of genes and retrotransposons at the barley Rar1 locus. Numbers above the black lines mark nt positions (kb) in the 66-kb contig. Gene exons are shown in blue. Retroelements are shown as color-coded thick lines, and arrows indicate sense direction of each element.
Figure 2
Retrotransposon BARE-1 insertions in the 66-kb contig. (A) Organization of a full-length 8.9-kb BARE-1 element as described in the text. The unit is drawn in the sense direction. (B) Organization of the BARE-1 domains on the contig. Arrows represent LTRs numbered as in the text. Black ovals (not to scale) indicate direct repeats found adjacent to the LTRs that were created upon BARE-1 insertion. (C) Proposed ancestral state of current genomic organization. Dotted lines indicate inter-/intrachromosomal recombination events resulting in loss of BARE-1 internal regions. The LTR-3 and -4 complex resulted from the insertion of one BARE-1 into another followed by recombination between two of the LTRs (shown by the arrow). LTR-5 and part of a UTL terminate the clone. (D) Model for recombination between two LTRs, resulting in a single recombinant LTR in the genome and a closed circle, bearing an LTR and the internal domain, which is then lost.
Figure 3
Sequence compilation of terminal LTR sequences. Terminal LTR sequences of BARE-1 (A), BAGY-2 (B), Sukkula (C), Sabrina (D), and Nikita (E) are boxed. Five–-base pair direct repeats flanking the LTRs are underlined. Numbering of LTRs is identical to Figure 1and indicated on the left of each sequence. GenBank accession numbers are given on the left for those LTRs that are not present in the 66-kb contig. Numbers above the DNA sequences refer to nt positions in the 66-kb contig or in other deposited GenBank sequences. Arrows indicate sense direction of each element.
Figure 4
Possible integration and recombination events generating Sabrina unit LTR-2 — LTR-3. Two Sabrina elements are shown in dark and bright gray with the arrows denoting the LTRs. Nonelement DNA is in black with the ovals representing 5-bp direct repeats flanking Sabrina LTR-2 and LTR-3. The broken arrow line shows a deduced intraelement recombination event; dotted lines indicate corresponding positions; thin black lines denote a nested Sabrina insertion.
Figure 5
Deduced nesting order of retroelements. Symbols and color codes correspond to Figure 1.
Comment on
- Are obese plant genomes on a diet?
Rabinowicz PD. Rabinowicz PD. Genome Res. 2000 Jul;10(7):893-4. doi: 10.1101/gr.10.7.893. Genome Res. 2000. PMID: 10899137 Review. No abstract available.
Similar articles
- Structural organization of the barley D-hordein locus in comparison with its orthologous regions of wheat genomes.
Gu YQ, Anderson OD, Londeorë CF, Kong X, Chibbar RN, Lazo GR. Gu YQ, et al. Genome. 2003 Dec;46(6):1084-97. doi: 10.1139/g03-071. Genome. 2003. PMID: 14663527 - Gene Deletion in Barley Mediated by LTR-retrotransposon BARE.
Shang Y, Yang F, Schulman AH, Zhu J, Jia Y, Wang J, Zhang XQ, Jia Q, Hua W, Yang J, Li C. Shang Y, et al. Sci Rep. 2017 Mar 2;7:43766. doi: 10.1038/srep43766. Sci Rep. 2017. PMID: 28252053 Free PMC article. - Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes.
Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH. Kalendar R, et al. Genetics. 2004 Mar;166(3):1437-50. doi: 10.1534/genetics.166.3.1437. Genetics. 2004. PMID: 15082561 Free PMC article. - LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model.
Vitte C, Panaud O. Vitte C, et al. Cytogenet Genome Res. 2005;110(1-4):91-107. doi: 10.1159/000084941. Cytogenet Genome Res. 2005. PMID: 16093661 Review. - A movable feast: diverse retrotransposons and their contribution to barley genome dynamics.
Schulman AH, Kalendar R. Schulman AH, et al. Cytogenet Genome Res. 2005;110(1-4):598-605. doi: 10.1159/000084993. Cytogenet Genome Res. 2005. PMID: 16093713 Review.
Cited by
- Small RNAs, DNA methylation and transposable elements in wheat.
Cantu D, Vanzetti LS, Sumner A, Dubcovsky M, Matvienko M, Distelfeld A, Michelmore RW, Dubcovsky J. Cantu D, et al. BMC Genomics. 2010 Jun 29;11:408. doi: 10.1186/1471-2164-11-408. BMC Genomics. 2010. PMID: 20584339 Free PMC article. - Transposable elements in a marginal plant population: temporal fluctuations provide new insights into genome evolution of wild diploid wheat.
Belyayev A, Kalendar R, Brodsky L, Nevo E, Schulman AH, Raskina O. Belyayev A, et al. Mob DNA. 2010 Feb 1;1(1):6. doi: 10.1186/1759-8753-1-6. Mob DNA. 2010. PMID: 20226076 Free PMC article. - Evolution of DNA amounts across land plants (embryophyta).
Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Leitch IJ, et al. Ann Bot. 2005 Jan;95(1):207-17. doi: 10.1093/aob/mci014. Ann Bot. 2005. PMID: 15596468 Free PMC article. - Demarcating the gene-rich regions of the wheat genome.
Erayman M, Sandhu D, Sidhu D, Dilbirligi M, Baenziger PS, Gill KS. Erayman M, et al. Nucleic Acids Res. 2004 Jul 7;32(12):3546-65. doi: 10.1093/nar/gkh639. Print 2004. Nucleic Acids Res. 2004. PMID: 15240829 Free PMC article. - The repetitive landscape of the chicken genome.
Wicker T, Robertson JS, Schulze SR, Feltus FA, Magrini V, Morrison JA, Mardis ER, Wilson RK, Peterson DG, Paterson AH, Ivarie R. Wicker T, et al. Genome Res. 2005 Jan;15(1):126-36. doi: 10.1101/gr.2438004. Epub 2004 Jul 15. Genome Res. 2005. PMID: 15256510 Free PMC article.
References
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms—583 new estimates. Ann Bot. 1997;80:169–196.
- Bennetzen JL, Schrick K, Springer PS, Brown WE, SanMiguel P. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. - PubMed
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources