Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences - PubMed (original) (raw)
Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences
D L Mager et al. J Virol. 2000 Aug.
Abstract
The repetitive ETn (early transposon) family of sequences represents an active "mobile mutagen" in the mouse genome. The presence of long terminal repeats (LTRs) and other diagnostic features indicate that ETns are retrotransposons but they contain no long open reading frames or documented similarity to the genes of known retroviruses or other retroelements. Thus, the mechanisms responsible for the mobility of this family have been unknown. In this study, we used computer searches to detect a small region of previously unrecognized type D retroviral pol homology within ETn elements. This small region was used to isolate two mouse endogenous proviral elements with gag, pro, and pol genes similar to simian type D viruses. This new family of mouse endogenous proviruses, termed MusD, is present in several hundred copies in the genome. Interestingly, the MusD LTRs, 3' internal region, and the 5' region expected to contain the packaging signal are very closely related to members of the ETn subfamily that have recently transposed. Analysis of different mouse strains indicates that MusD elements predate the existence of the mobile subfamily of ETns. These findings indicate that the ETn family was likely created via recombination events resulting in a near complete substitution of MusD coding sequences with unrelated DNA. Furthermore, these results suggest that ETn transcripts retrotranspose using proteins provided by MusD proviruses.
Figures
FIG. 1
Similarity between ETn elements and primate type D retroviruses. The amino acid translation of nucleotides 3618 to 3764 of the ETn element at the tyrosinase locus (10) is compared to the 3′ end of the pol protein of MPMV. Identical residues are in bold.
FIG. 2
Representation of three MusD elements. Thick lines are MusD sequences, and the filled boxes indicate the LTRs. The location of the EST AA142642 is shown as a thin line, and locations of the PCR primers used to amplify the region deleted in AE000665 are shown as small arrows. The gag, pro, and pol genes are represented as ovals, with the ORF-destroying mutations shown as asterisks (for single-nucleotide-length differences or substitutions) and triangles (for the 4- and 14-bp deletions described in the text).
FIG. 3
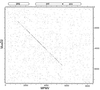
Dot matrix nucleotide comparison of MusD2 and MPMV. Extents of the MPMV genes are indicated. The stringency of comparison was 15 out of 23.
FIG. 4
(A) DNA sequence of the 5′ LTR and 5′ internal region of MusD1. The LTR is outlined, and potential TATAA and polyadenylation signals are boxed. The putative Lys-tRNA primer binding site is underlined, and the region forming the stem-loop structure shown in panel B is overlined. The gag initiation codon is shown with a double underline. (B) Potential stem-loop structure with the ACC motif present in the loop.
FIG. 5
Amino acid comparisons of MusD to MPMV. (A) Comparison of the translated MusD1 sequence to the MPMV gag products. MPMV p10, pp24, and p12 span residues 1 to 299 and p27, p14, and p4 span residues 300 to 657. The location of the 14-bp insertion added to maintain the MusD1 reading frame is indicated (residues 266 to 270 of MusD1). The major homology region is underlined, and the highly conserved residues are shown with filled circles. The conserved cysteine and histidine residues in the two zinc finger motifs are indicated with a filled triangle. (B) Comparison of the translated MusD2 sequence to the MPMV pro product. The enzymatic active site, the “flap” region, and the GRDLL conserved domains are shown by an arrowed line, a solid line, and a dashed line, respectively. (C) Comparison of the translated MusD2 sequence to the MPMV pol product. The four positions which were corrected based on other MusD sequences to maintain the ORF are indicated with a slanted line through the sequence. The highly conserved residues in the reverse transcriptase, the RNase H, and the integrase domains discussed in the text are indicated by filled circles, asterisks, and triangles, respectively.
FIG. 6
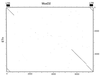
Dot matrix nucleotide comparison of MusD2 and the ETn element at the tyrosinase locus (10). The stringency of comparison was 17 out of 23.
FIG. 7
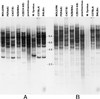
(A) Genomic Southern analysis of _Eco_RI-digested DNAs from different mouse strains by using a 250-bp MusD-specific probe (25-h exposure). (B) Rehybridization of the same blot with a 24-mer oligonucleotide probe specific for the type 2 ETn subfamily (10-day exposure).
FIG. 8
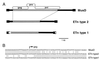
(A) Representation of the relationship between MusD elements and ETns. The vertically striped region shows MusD-specific sequences. Diagonally striped boxes show ETn-specific sequences, and white boxes show regions specific to the type 1 ETn subfamily. (B) Nucleotide sequence of the three element types at the start of the gag gene. MusD is the MusD1 sequence, ETn type 2 is the tyrosinase locus element (10), and ETn type 1 is GenBank accession no. M16478 (26).
Similar articles
- An active murine transposon family pair: retrotransposition of "master" MusD copies and ETn trans-mobilization.
Ribet D, Dewannieux M, Heidmann T. Ribet D, et al. Genome Res. 2004 Nov;14(11):2261-7. doi: 10.1101/gr.2924904. Epub 2004 Oct 12. Genome Res. 2004. PMID: 15479948 Free PMC article. - Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse.
Baust C, Gagnier L, Baillie GJ, Harris MJ, Juriloff DM, Mager DL. Baust C, et al. J Virol. 2003 Nov;77(21):11448-58. doi: 10.1128/jvi.77.21.11448-11458.2003. J Virol. 2003. PMID: 14557630 Free PMC article. - Insertional polymorphisms of ETn retrotransposons include a disruption of the wiz gene in C57BL/6 mice.
Baust C, Baillie GJ, Mager DL. Baust C, et al. Mamm Genome. 2002 Aug;13(8):423-8. doi: 10.1007/s00335-002-2178-3. Mamm Genome. 2002. PMID: 12226707 - Molecular biology of type A endogenous retrovirus.
Ono M. Ono M. Kitasato Arch Exp Med. 1990 Sep;63(2-3):77-90. Kitasato Arch Exp Med. 1990. PMID: 1710682 Review. - Endogenous retroviruses: still active after all these years?
Stoye JP. Stoye JP. Curr Biol. 2001 Nov 13;11(22):R914-6. doi: 10.1016/s0960-9822(01)00553-x. Curr Biol. 2001. PMID: 11719237 Review.
Cited by
- Next-generation sequencing identifies the Danforth's short tail mouse mutation as a retrotransposon insertion affecting Ptf1a expression.
Vlangos CN, Siuniak AN, Robinson D, Chinnaiyan AM, Lyons RH Jr, Cavalcoli JD, Keegan CE. Vlangos CN, et al. PLoS Genet. 2013;9(2):e1003205. doi: 10.1371/journal.pgen.1003205. Epub 2013 Feb 21. PLoS Genet. 2013. PMID: 23437000 Free PMC article. - Evolution and distribution of class II-related endogenous retroviruses.
Gifford R, Kabat P, Martin J, Lynch C, Tristem M. Gifford R, et al. J Virol. 2005 May;79(10):6478-86. doi: 10.1128/JVI.79.10.6478-6486.2005. J Virol. 2005. PMID: 15858031 Free PMC article. - Identification, phylogeny, and evolution of retroviral elements based on their envelope genes.
Bénit L, Dessen P, Heidmann T. Bénit L, et al. J Virol. 2001 Dec;75(23):11709-19. doi: 10.1128/JVI.75.23.11709-11719.2001. J Virol. 2001. PMID: 11689652 Free PMC article. - A novel active endogenous retrovirus family contributes to genome variability in rat inbred strains.
Wang Y, Liska F, Gosele C, Sedová L, Kren V, Krenová D, Ivics Z, Hubner N, Izsvák Z. Wang Y, et al. Genome Res. 2010 Jan;20(1):19-27. doi: 10.1101/gr.100073.109. Epub 2009 Nov 3. Genome Res. 2010. PMID: 19887576 Free PMC article. - RNase H2, mutated in Aicardi-Goutières syndrome, promotes LINE-1 retrotransposition.
Benitez-Guijarro M, Lopez-Ruiz C, Tarnauskaitė Ž, Murina O, Mian Mohammad M, Williams TC, Fluteau A, Sanchez L, Vilar-Astasio R, Garcia-Canadas M, Cano D, Kempen MH, Sanchez-Pozo A, Heras SR, Jackson AP, Reijns MA, Garcia-Perez JL. Benitez-Guijarro M, et al. EMBO J. 2018 Aug 1;37(15):e98506. doi: 10.15252/embj.201798506. Epub 2018 Jun 29. EMBO J. 2018. PMID: 29959219 Free PMC article.
References
- Beck J A, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig J T, Festing M F, Fisher E M. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases