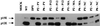Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses - PubMed (original) (raw)
Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses
B Yuan et al. J Virol. 2000 Aug.
Abstract
The p12 region of the Moloney murine leukemia virus (M-MuLV) Gag protein contains a PPPY motif important for efficient virion assembly and release. To probe the function of the PPPY motif, a series of insertions of homologous and heterologous motifs from other retroviruses were introduced at various positions in a mutant gag gene lacking the PPPY motif. The assembly defects of the PPPY deletion mutant could be rescued by insertion of a wild-type PPPY motif and flanking sequences at several ectopic positions in the Gag protein. The late assembly domain (L-domain) of Rous sarcoma virus (RSV) or human immunodeficiency virus type 1 (HIV-1) could also fully or partially restore M-MuLV assembly when introduced into matrix, p12, or nucleocapsid domains of the mutant M-MuLV Gag protein lacking the PPPY motif. Strikingly, mutant viruses carrying the RSV or the HIV-1 L-domain at the original location of the deleted PPPY motif were replication competent in rodent cells. These data suggest that the PPPY motif of M-MuLV acts in a partially position-independent manner and is functionally interchangeable with L-domains of other retroviruses. Electron microscopy studies revealed that deletion of the entire p12 region resulted in the formation of tube-like rather than spherical particles. Remarkably, the PPPY deletion mutant formed chain structures composed of multiple viral particles linked on the cell surface. Many of the mutants with heterologous L-domains released virions with wild-type morphology.
Figures
FIG. 1
Mutations in the M-MuLV Gag polyprotein. The M-MuLV Gag protein is represented by rectangular boxes. The location of the PPPY motif in the p12 region of the wild-type Gag protein is indicated. The entire p12 region is deleted in the Del-p12 mutant. The DPY mutant contains a 5-amino acid deletion (DPPPY) which removes the PPPY motif; this mutant was used as the parent to generate the insertion mutants. Three different fragments were used for the insertion mutants: PY, an 11-amino-acid fragment containing the wild-type PPPY motif and flanking sequences from M-MuLV; p2b, the entire RSV p2b sequence (11 amino acids) including the PPPPY motif; and P6, the N-terminal 18 amino acids of the HIV-1 p6 protein including the PTAPP motif. The amino acid (aa) sequences of PY, p2b, and P6 are as shown. The insertions are represented by black boxes. The locations of insertions are as indicated, with the names of the mutants on the left.
FIG. 2
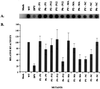
Viral assembly and particle release analyzed by the RT assay. (A) RT activity in the culture supernatant. The proviruses of the insertion mutants were transiently transfected into 293T cells by calcium phosphate precipitation, and supernatants were analyzed by the RT assay 48 h after transfection. (B) PhosphorImager quantitation of the relative RT activity in produced virions. The mean of three independent experiments is shown. Error bars indicate standard deviation of the three results. The amount of RT activity in the wild-type (WT) virus is set as 100.
FIG. 3
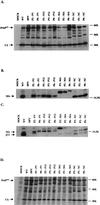
Western blot analysis of gag gene products in virions and in lysates of transfected cells. The virus particles were collected by ultracentrifugation from the culture supernatants of the transfected 293T cells. The virions were lysed, and proteins were subjected to Western blot analysis. Blots were probed with polyclonal antisera against CA (A), MA (B), and p12 (C). The anti-p12 antiserum also showed contaminating reactivity to MA. (D) Lysates of transfected 293T cells were also analyzed by Western blotting using anti-CA polyclonal serum. The positions of Pr65_gag_, CA, MA, and p12 are indicated. WT, wild type.
FIG. 4
Analysis of TM envelope protein in mutant virions. Virions were harvested, and proteins were analyzed by Western blotting. A TM-specific monoclonal antiserum was used to detect p15E and p12E forms of the TM protein. The positions of p15E and p12E are indicated. WT, wild type.
FIG. 5
Southern blot detection of linear and circular viral DNAs synthesized in infected NIH 3T3 cells. Supernatants from transfected 293T cells were used to acutely infect NIH 3T3 cells. The virus titers in the supernatants were normalized for RT activity. Low-molecular-weight DNAs were extracted by the Hirt method, and the viral DNAs were detected by hybridization with a radiolabeled virus-specific DNA probe. The same membrane was stripped and rehybridized with a mitochondrial DNA probe to monitor the recovery of DNA loaded in each lane. The positions and sizes of linear and two circular viral DNAs are marked. WT, wild type.
FIG. 6
Virus infectivity. Equal volumes of culture medium from transfected 293T cells were collected 48 h after transfection and used to infect naive NIH 3T3 cells. Culture supernatants were harvested every day up to 17 days, and the virus yield was monitored by the RT assay. The RT assay results from day 2 to day 9 are shown. The days when cultures were split are marked. WT, wild type.
FIG. 7
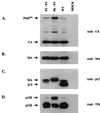
Western blot analysis of virion proteins in the replication-competent chimeric mutants. The culture supernatants of infected NIH 3T3 cells in Fig. 6 were collected on day 6. The viral particles were pelleted by ultracentrifugation, lysed, and analyzed by Western blotting. The membranes were probed with anti-CA (A), anti-MA (B), anti-p12 (C) polyclonal antiserum. (D) In addition, a specific monoclonal antibody was used to detect TM envelope protein. The anti-p12 polyclonal antiserum also showed contaminating reactivity with MA. The positions of Pr65_gag_, CA, MA, and p12 are indicated. WT, wild type.
FIG. 8
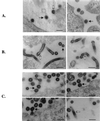
EM images of 293T cells expressing wild-type and mutant viral genomes. (A) Wild-type virus particle release. The mature viruses are marked by black arrows, and the immature viruses are marked by white arrows. (B) Particle release from the Del-p12 mutant. The tube-like structures are indicated by white arrows. A black arrow points to a cross-section of a tube. (C) Particle release from the DPY mutant. The multiple virus chain structures are marked by black arrows, and tubes are indicated by white arrows. Bars, 200 nm.
FIG. 9
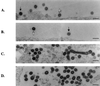
EM images of virions released from 293T cells transfected by the insertion mutants. (A) Mutant P2-PY. (B) Mutant P6-PY. In these panels, black arrows point to mature virions and white arrows point to immature virions (C) Mutant P6-P12. (D) Mutant P6-MA. In these two panels, the chain structures are indicated by white arrows and the tube structures are indicated by black arrows. Scale bars, 200 nm.
Similar articles
- Late domain-independent rescue of a release-deficient Moloney murine leukemia virus by the ubiquitin ligase itch.
Jadwin JA, Rudd V, Sette P, Challa S, Bouamr F. Jadwin JA, et al. J Virol. 2010 Jan;84(2):704-15. doi: 10.1128/JVI.01319-09. Epub 2009 Oct 28. J Virol. 2010. PMID: 19864377 Free PMC article. - Repression of the Chromatin-Tethering Domain of Murine Leukemia Virus p12.
Brzezinski JD, Modi A, Liu M, Roth MJ. Brzezinski JD, et al. J Virol. 2016 Nov 28;90(24):11197-11207. doi: 10.1128/JVI.01084-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707926 Free PMC article. - Phosphorylation Requirement of Murine Leukemia Virus p12.
Brzezinski JD, Felkner R, Modi A, Liu M, Roth MJ. Brzezinski JD, et al. J Virol. 2016 Nov 28;90(24):11208-11219. doi: 10.1128/JVI.01178-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707931 Free PMC article. - Particle assembly and genome packaging.
Linial ML, Eastman SW. Linial ML, et al. Curr Top Microbiol Immunol. 2003;277:89-110. doi: 10.1007/978-3-642-55701-9_4. Curr Top Microbiol Immunol. 2003. PMID: 12908769 Review. - Mutational analysis of HIV-1 gag proteins (review).
Miyaura M, Yoshida A, Sakurai A, Fujita M, Koyama AH, Adachi A. Miyaura M, et al. Int J Mol Med. 2000 Sep;6(3):265-9. doi: 10.3892/ijmm.6.3.265. Int J Mol Med. 2000. PMID: 10934287 Review.
Cited by
- Viral DNA tethering domains complement replication-defective mutations in the p12 protein of MuLV Gag.
Schneider WM, Brzezinski JD, Aiyer S, Malani N, Gyuricza M, Bushman FD, Roth MJ. Schneider WM, et al. Proc Natl Acad Sci U S A. 2013 Jun 4;110(23):9487-92. doi: 10.1073/pnas.1221736110. Epub 2013 May 9. Proc Natl Acad Sci U S A. 2013. PMID: 23661057 Free PMC article. - Murine leukemia viruses: objects and organisms.
Rein A. Rein A. Adv Virol. 2011;2011:403419. doi: 10.1155/2011/403419. Epub 2011 Nov 15. Adv Virol. 2011. PMID: 22312342 Free PMC article. - YRKL sequence of influenza virus M1 functions as the L domain motif and interacts with VPS28 and Cdc42.
Hui EK, Barman S, Tang DH, France B, Nayak DP. Hui EK, et al. J Virol. 2006 Mar;80(5):2291-308. doi: 10.1128/JVI.80.5.2291-2308.2006. J Virol. 2006. PMID: 16474136 Free PMC article. Retracted. - Nucleic acid-independent retrovirus assembly can be driven by dimerization.
Johnson MC, Scobie HM, Ma YM, Vogt VM. Johnson MC, et al. J Virol. 2002 Nov;76(22):11177-85. doi: 10.1128/jvi.76.22.11177-11185.2002. J Virol. 2002. PMID: 12388677 Free PMC article. - Context-dependent effects of L domains and ubiquitination on viral budding.
Martin-Serrano J, Perez-Caballero D, Bieniasz PD. Martin-Serrano J, et al. J Virol. 2004 Jun;78(11):5554-63. doi: 10.1128/JVI.78.11.5554-5563.2004. J Virol. 2004. PMID: 15140952 Free PMC article.
References
- Arcement L, Karshin W, Naso R, Jamjoom G, Arlinghaus R. Biosynthesis of Rauscher leukemia viral proteins: presence of p30 and envelope p15 sequences in precursor polypeptides. Virology. 1976;69:763–774. - PubMed
- Arcement L J, Karshin W I, Naso R B, Arlinghaus R B. “Gag” polyprotein precursors of Rauscher murine leukemia virus. Virology. 1977;81:284–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials