Genetic inactivation of an inwardly rectifying potassium channel (Kir4.1 subunit) in mice: phenotypic impact in retina - PubMed (original) (raw)
Genetic inactivation of an inwardly rectifying potassium channel (Kir4.1 subunit) in mice: phenotypic impact in retina
P Kofuji et al. J Neurosci. 2000.
Abstract
The inwardly rectifying potassium channel Kir4.1 has been suggested to underlie the principal K(+) conductance of mammalian Müller cells and to participate in the generation of field potentials and regulation of extracellular K(+) in the retina. To further assess the role of Kir4.1 in the retina, we generated a mouse line with targeted disruption of the Kir4.1 gene (Kir4.1 -/-). Müller cells from Kir4.1 -/- mice were not labeled with an anti-Kir4.1 antibody, although they appeared morphologically normal when stained with an anti-glutamine synthetase antibody. In contrast, in Müller cells from wild-type littermate (Kir4.1 +/+) mice, Kir4.1 was present and localized to the proximal endfeet and perivascular processes. In situ whole-cell patch-clamp recordings showed a 10-fold increase in the input resistance and a large depolarization of Kir4.1 -/- Müller cells compared with Kir4.1 +/+ cells. The slow PIII response of the light-evoked electroretinogram (ERG), which is generated by K(+) fluxes through Müller cells, was totally absent in retinas from Kir4.1 -/- mice. The b-wave of the ERG, in contrast, was spared in the null mice. Overall, these results indicate that Kir4.1 is the principal K(+) channel subunit expressed in mouse Müller glial cells. The highly regulated localization and the functional properties of Kir4.1 in Müller cells suggest the involvement of this channel in the regulation of extracellular K(+) in the mouse retina.
Figures
Fig. 1.
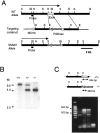
Targeted strategy for Kir4.1 gene interruption. A, Part of the wild-type Kir4.1 locus containing the coding exon (WT Allele), the targeting construct, and the targeted locus (Mutant Allele) are shown. PGKneo, Neomycin resistance gene.MC1tk, Thymidine kinase gene. The sizes of the_Xho_I fragments predicted to hybridize to the indicated diagnostic probe are shown. Restriction endonucleases:B, _Bam_HI; Bl,_Bgl_II; E, _Eco_RI;H, _Hin_dIII; S,_Sac_I; X, _Xho_I.B, Southern blot analysis of_Xho_I-digested genomic DNA from wild-type (+/+), heterozygous (+/−), and homozygous mutant mice (−/−) with the diagnostic probe indicated in A. C, PCR analysis of tail genomic DNA PCR primers amplify a 634 bp fragment in the +/+ allele and a 383 bp fragment in the mutant allele. Notice the 634 bp fragment obtained from PCR analysis using tail DNA from +/+ mouse, the 383 bp fragment using tail DNA from −/− mouse, and both fragments from +/− mouse. As a control (C), the PCR analysis was performed in the absence of mouse genomic DNA
Fig. 2.
Analysis of Kir4.1 mRNA in retina. PCR analysis of total RNA extracted from P21 Kir4.1 +/+ and Kir4.1 −/− mice retinas and Kir4.1 +/+ brain. Kir-specific oligonucleotide pairs were used to determine the expression of the various Kir channels subunits in retina and brain. As a control (C), cDNA was replaced by water, and the PCR was performed using Kir5.1-specific oligonucleotide primers.
Fig. 3.
Histological analyses of the retinas of the Kir4.1 +/+ and Kir4.1 −/− mice. Cross-sections of mouse retinas from +/+ and −/− mutant mice (P11) were stained with hematoxylin and eosin.GCL, Ganglion cell layer; OPL, outer plexiform layer. Scale bar, 25 μm.
Fig. 4.
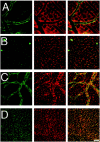
Immunohistochemistry of Kir4.1 in retinal whole mounts from Kir4.1 +/+ mouse (P18). Retinal whole mount stained with anti-Kir4.1 antibody (green) and anti-GS antibody (red). Confocal images were obtained in the ganglion cell layer (A), inner plexiform layer (B), inner nuclear layer (C), and outer nuclear layer (D). Note that the expression of Kir4.1 was clustered around neurons in the outer nuclear layer and along the blood vessels in the superficial and inner nuclear layer. Immunofluorescence in the blood vessels revealed by Texas Red donkey anti-mouse antibody (C) represents the binding of the secondary antibody to mouse IgG, because this immunoreactivity is not seen in retinas from mice transcardially perfused with PBS to wash out the blood before fixation. Scale bar, 20 μm.
Fig. 5.
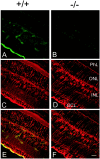
Immunohistochemical analysis of Kir4.1 in retinal sections of Kir4.1 +/+ (+/+) (A,C, E) and Kir4.1 −/− (−/−) (B, D, F) P18 mice. Sections were double-stained with affinity-purified rabbit anti-rat Kir4.1 antibody, followed by FITC-conjugated anti-rabbit IgG (A, B, green), and monoclonal anti-GS antibody, followed by Texas Red-conjugated anti-mouse IgG (C, D,red). E, F, Superposition of images A, C and B,D, respectively. Scale bar, 20 μm. GCL, Ganglion cell layer; PhL, photoreceptor layer.
Fig. 6.
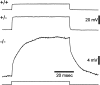
Input resistance of Müller cells in Kir4.1 +/+, Kir4.1 +/−, and Kir4.1 −/− mice. Traces show the displacement of the membrane potential produced by an injected current pulse. The traces are scaled so that the amplitudes of the responses reflect the relative input resistances of the three cells (voltage divided by injected current is equal for all_traces_). Note that the membrane time constant of the −/− cell is substantially longer because of its increased membrane resistance. Amplitude of current pulses: 0.5 nA for +/+ and +/−; 0.1 nA for −/−.
Fig. 7.
The slow PIII response of the ERG in Kir4.1 +/+ and Kir4.1 −/− mice. Traces show the intraretinal ERG recorded between an electrode in the distal retina and one in the vitreous humor. In a Kir4.1 +/+ mouse, a transient negative b-wave and a slower positive slow PIII response are evident (+/+). When Ba2+ (0.4 m
m
) is added to the superfusate (+/+ plus Ba 2+), the slow PIII response is abolished, whereas the b-wave remains primarily unchanged. Subtracting the trace in Ba2+from the other (+/+ difference) reveals the Ba2+-sensitive component of the ERG, the slow PIII response in isolation. In a Kir4.1 −/− mouse (−/−), the b-wave is present but the slow PIII response is absent. Addition of Ba2+ (−/− plus Ba2+) produces a small increase in the b-wave but no change in the response after decay of the b-wave, as illustrated by the difference_trace_ (−/− difference). The time course of the light stimulus is shown at the bottom.Dashed lines indicate prestimulus baseline levels.
Similar articles
- Dystrophin Dp71 is critical for the clustered localization of potassium channels in retinal glial cells.
Connors NC, Kofuji P. Connors NC, et al. J Neurosci. 2002 Jun 1;22(11):4321-7. doi: 10.1523/JNEUROSCI.22-11-04321.2002. J Neurosci. 2002. PMID: 12040037 Free PMC article. - Expression and clustered distribution of an inwardly rectifying potassium channel, KAB-2/Kir4.1, on mammalian retinal Müller cell membrane: their regulation by insulin and laminin signals.
Ishii M, Horio Y, Tada Y, Hibino H, Inanobe A, Ito M, Yamada M, Gotow T, Uchiyama Y, Kurachi Y. Ishii M, et al. J Neurosci. 1997 Oct 15;17(20):7725-35. doi: 10.1523/JNEUROSCI.17-20-07725.1997. J Neurosci. 1997. PMID: 9315894 Free PMC article. - Inwardly rectifying K+ channel in retinal Müller cells: comparison with the KAB-2/Kir4.1 channel expressed in HEK293T cells.
Tada Y, Horio Y, Kurachi Y. Tada Y, et al. Jpn J Physiol. 1998 Feb;48(1):71-80. doi: 10.2170/jjphysiol.48.71. Jpn J Physiol. 1998. PMID: 9538292 - Role of glial K(+) channels in ontogeny and gliosis: a hypothesis based upon studies on Müller cells.
Bringmann A, Francke M, Pannicke T, Biedermann B, Kodal H, Faude F, Reichelt W, Reichenbach A. Bringmann A, et al. Glia. 2000 Jan 1;29(1):35-44. doi: 10.1002/(sici)1098-1136(20000101)29:1<35::aid-glia4>3.0.co;2-a. Glia. 2000. PMID: 10594921 Review. - Molecular substrates of potassium spatial buffering in glial cells.
Kofuji P, Connors NC. Kofuji P, et al. Mol Neurobiol. 2003 Oct;28(2):195-208. doi: 10.1385/MN:28:2:195. Mol Neurobiol. 2003. PMID: 14576456 Review.
Cited by
- Astrocytic glutamate uptake is slow and does not limit neuronal NMDA receptor activation in the neonatal neocortex.
Hanson E, Armbruster M, Cantu D, Andresen L, Taylor A, Danbolt NC, Dulla CG. Hanson E, et al. Glia. 2015 Oct;63(10):1784-96. doi: 10.1002/glia.22844. Epub 2015 Apr 27. Glia. 2015. PMID: 25914127 Free PMC article. - Dystrophin Dp71: the smallest but multifunctional product of the Duchenne muscular dystrophy gene.
Tadayoni R, Rendon A, Soria-Jasso LE, Cisneros B. Tadayoni R, et al. Mol Neurobiol. 2012 Feb;45(1):43-60. doi: 10.1007/s12035-011-8218-9. Epub 2011 Nov 22. Mol Neurobiol. 2012. PMID: 22105192 Review. - Cannabinoid Receptors CB1 and CB2 Modulate the Electroretinographic Waves in Vervet Monkeys.
Bouskila J, Harrar V, Javadi P, Beierschmitt A, Palmour R, Casanova C, Bouchard JF, Ptito M. Bouskila J, et al. Neural Plast. 2016;2016:1253245. doi: 10.1155/2016/1253245. Epub 2016 Mar 16. Neural Plast. 2016. PMID: 27069692 Free PMC article. - A silk platform that enables electrophysiology and targeted drug delivery in brain astroglial cells.
Benfenati V, Toffanin S, Capelli R, Camassa LM, Ferroni S, Kaplan DL, Omenetto FG, Muccini M, Zamboni R. Benfenati V, et al. Biomaterials. 2010 Nov;31(31):7883-91. doi: 10.1016/j.biomaterials.2010.07.013. Epub 2010 Aug 4. Biomaterials. 2010. PMID: 20688390 Free PMC article. - Prph2 disease mutations lead to structural and functional defects in the RPE.
Tebbe L, Sakthivel H, Makia MS, Kakakhel M, Conley SM, Al-Ubaidi MR, Naash MI. Tebbe L, et al. FASEB J. 2022 May;36(5):e22284. doi: 10.1096/fj.202101562RR. FASEB J. 2022. PMID: 35344225 Free PMC article.
References
- Ando M, Takeuchi S. Immunological identification of an inward rectifier K+ channel (Kir4.1) in the intermediate cell (melanocyte) of the cochlear stria vascularis of gerbils and rats. Cell Tissue Res. 1999;298:179–183. - PubMed
- Bignami A, Dahl D. The radial glia of Müller in the rat retina and their response to injury. An immunofluorescence study with antibodies to the glial fibrillary acidic (GFA) protein. Exp Eye Res. 1979;28:63–69. - PubMed
- Bolnick DA, Walter AE, Sillman AJ. Barium suppresses slow PIII in perfused bullfrog retina. Vision Res. 1979;19:1117–1119. - PubMed
- Bond C, Pessia M, Xia X, Lagrutta A, Kavanaugh M, Adelman J. Cloning and expression of a family of inward rectifier potassium channels. Receptors Channels. 1994;2:183–191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MH-49176/MH/NIMH NIH HHS/United States
- R01 EY012949/EY/NEI NIH HHS/United States
- GM-29836/GM/NIGMS NIH HHS/United States
- R01 EY004077/EY/NEI NIH HHS/United States
- EY04077/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases