A carboxy-terminal 16-amino-acid region of sigma(38) of Escherichia coli is important for transcription under high-salt conditions and sigma activities in vivo - PubMed (original) (raw)
A carboxy-terminal 16-amino-acid region of sigma(38) of Escherichia coli is important for transcription under high-salt conditions and sigma activities in vivo
M Ohnuma et al. J Bacteriol. 2000 Aug.
Abstract
sigma(38) (or sigma(S), the rpoS gene product) is a sigma subunit of RNA polymerase in Escherichia coli and directs transcription from a number of stationary-phase promoters as well as osmotically inducible promoters. In this study, we analyzed the function of the carboxy-terminal 16-amino-acid region of sigma(38) (residues 315 to 330), which is well conserved among the rpoS gene products of enteric bacterial species. Truncation of this region was shown to result in the loss of sigma activity in vivo using promoter-lacZ fusion constructs, but the mutant sigma(38) retained the binding activity in vivo to the core enzyme. The in vitro transcription analysis revealed that the transcription activity of sigma(38) holoenzyme under high potassium glutamate concentrations was significantly decreased by the truncation of the carboxy-terminal tail element.
Figures
FIG. 1
Structure of the carboxy-terminal region of RpoS. The thick bar represents a linear diagram of ς38. N and C indicate the amino and carboxy termini of ς38, respectively. Conserved regions 1 to 4 are indicated, and subregions 1.1, 2.1, 2.4, and 4.2, for which functions were assigned, are indicated above the thick bar. Amino acid sequences of the carboxy-terminal region are aligned below the thick bar. GenBank database searches were performed by the BLAST program through homepages of National Center for Biotechnology Information (
). Amino acid sequences were aligned using the Clustal V program with a fixed gap penalty of 10 and a floating gap penalty of 10. Asterisks represent identical residues, and periods represent residues having similar characteristics. The abbreviations denote the following bacterial species: Eco, E. coli; Ecl, Enterobacter cloacae; Eca, Erwinia carotovora; Pae, Pseudomonas aeruginosa; Pfl, Pseudomonas fluorescens; Ppu, Pseudomonas putida; Sente, Salmonella enterica; Sty, S. enterica serovar Typhimurium; Sento, Serratia entomophila; and Yen, Yersinia enterocolitica. A bracket indicates a helix-turn-helix DNA binding motif at region 4.2. The shaded area indicates the carboxy-terminal 16-amino-acid region (CTE) that was truncated in this study.
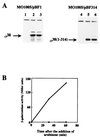
FIG. 2
Effect of truncation of ς38-CTE on the transcription activity of the katE promoter. MO1005EL cells harboring pBF1 or pBF314 were grown with shaking in Luria-Bertani medium supplemented with kanamycin (50 μg/ml), tetracycline (10 μg/ml), and ampicillin (50 μl/ml) at 37°C. RpoS proteins were induced by the addition of arabinose at an optical density at 660 nm of 0.3. (A) The expression of ς38 or ς38(1-314) was monitored by Western analysis with antiserum against ς38 (25). Samples were taken at 0 min (lanes 1 and 4), 30 min (lanes 2 and 5), and 60 min (lanes 3 and 6) after the addition of arabinose. The upper signals are an unrelated band detected by the serum that are irrespective of the rpoS expression. (B) The katE transcription activity was assayed using a lacZ fusion construct. The β-galactosidase activity was assayed as previously described (5) except that cell density was measured at 660 nm. The solid line and dashed line correspond to results obtained with MO1005EL/pBF1 and MO1005EL/pBF314, respectively. These results are averages of duplicate measurements.
FIG. 3
ς38-CTE is not required for holoenzyme formation. pTZ19R (vector) (A), pKTF1 (ς38 overexpression plasmid) (B), and pKTF314 [ς38(1-314) overexpression plasmid] (C) were introduced into rpoS null mutant MO1001FL. Effects on growth and fic-lacZ expression resulted from ς38, and ς38(1-314) overexpression was monitored using a Luria-Bertani plate containing 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-
d
-galactopyranoside)/ml and 0.1 mM IPTG (isopropyl-β-
d
-thiogalactopyranoside).
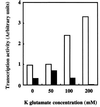
FIG. 4
ς38-CTE is required for effective transcription activity at high ionic strength in vitro. The core enzyme and either ς38 or ς38(1-314) were mixed at the ratio of 1:4 and incubated for 30 min at 37°C to reconstitute holoenzymes. Single-round in vitro transcription assays were performed essentially as previously described (17). Transcription reactions were carried out under various ionic strengths (0, 50, 100, and 200 mM of K glutamate) with 100 nM reconstituted holoenzyme and 3 nM template DNA containing the fic promoter, i.e., the _Bam_HI-_Eco_RI fragment (776 bp) of pFL1 (26). The DNA template produced in vitro transcripts 242 nucleotides in length. Transcription activity at 50 mM K glutamate by Eς38 was set at 1. Open bars represent transcription activity of Eς38, and closed bars represent that of Eς38(1-314). These results are averages of duplicate experiments.
Similar articles
- Recognition of promoter DNA by subdomain 4.2 of Escherichia coli sigma 70: a knowledge based model of -35 hexamer interaction with 4.2 helix-turn-helix motif.
Reddy BV, Gopal V, Chatterji D. Reddy BV, et al. J Biomol Struct Dyn. 1997 Feb;14(4):407-19. doi: 10.1080/07391102.1997.10508140. J Biomol Struct Dyn. 1997. PMID: 9172641 - Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli.
Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Tanaka K, et al. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3511-5. doi: 10.1073/pnas.90.8.3511. Proc Natl Acad Sci U S A. 1993. PMID: 8475100 Free PMC article. - Interaction of Escherichia coli sigma 70 with core RNA polymerase.
Burgess RR, Arthur TM, Pietz BC. Burgess RR, et al. Cold Spring Harb Symp Quant Biol. 1998;63:277-87. doi: 10.1101/sqb.1998.63.277. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384292 Review. No abstract available. - How sigma docks to RNA polymerase and what sigma does.
Burgess RR, Anthony L. Burgess RR, et al. Curr Opin Microbiol. 2001 Apr;4(2):126-31. doi: 10.1016/s1369-5274(00)00177-6. Curr Opin Microbiol. 2001. PMID: 11282466 Review.
Cited by
- In vitro properties of RpoS (sigma(S)) mutants of Escherichia coli with postulated N-terminal subregion 1.1 or C-terminal region 4 deleted.
Gowrishankar J, Yamamoto K, Subbarayan PR, Ishihama A. Gowrishankar J, et al. J Bacteriol. 2003 Apr;185(8):2673-9. doi: 10.1128/JB.185.8.2673-2679.2003. J Bacteriol. 2003. PMID: 12670993 Free PMC article. - A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli.
Cheung KJ, Badarinarayana V, Selinger DW, Janse D, Church GM. Cheung KJ, et al. Genome Res. 2003 Feb;13(2):206-15. doi: 10.1101/gr.401003. Genome Res. 2003. PMID: 12566398 Free PMC article. - A comparative study of variation in codon 33 of the rpoS gene in Escherichia coli K12 stocks: implications for the synthesis of sigma(s).
Subbarayan PR, Sarkar M. Subbarayan PR, et al. Mol Genet Genomics. 2004 Jan;270(6):533-8. doi: 10.1007/s00438-003-0944-x. Epub 2003 Nov 14. Mol Genet Genomics. 2004. PMID: 14618393 - Poising of Escherichia coli RNA polymerase and its release from the sigma 38 C-terminal tail for osmY transcription.
Rosenthal AZ, Kim Y, Gralla JD. Rosenthal AZ, et al. J Mol Biol. 2008 Feb 29;376(4):938-49. doi: 10.1016/j.jmb.2007.12.037. Epub 2008 Jan 16. J Mol Biol. 2008. PMID: 18201723 Free PMC article. - Regulation of sigma factor competition by the alarmone ppGpp.
Jishage M, Kvint K, Shingler V, Nyström T. Jishage M, et al. Genes Dev. 2002 May 15;16(10):1260-70. doi: 10.1101/gad.227902. Genes Dev. 2002. PMID: 12023304 Free PMC article.
References
- Ding Q, Kusano S, Villarejo M, Ishihama A. Promoter selectivity control of Escherichia coli RNA polymerase by ionic strength: differential recognition of osmoregulated promoters by EςD and EςS holoenzymes. Mol Microbiol. 1995;16:649–656. - PubMed
- Dombroski A J, Walter W A, Record M T, Jr, Siegele D A, Gross C A. Polypeptides containing highly conserved regions of transcription factor ς70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. - PubMed
- Espinosa-Urgel M, Chamizo C, Tormo A. A consensus structure for ςS-dependent promoters. Mol Microbiol. 1996;21:657–659. - PubMed
- Farewell A, Kvint K, Nyström T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998;29:1039–1051. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources