The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p - PubMed (original) (raw)
The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p
C Bonnerot et al. Mol Cell Biol. 2000 Aug.
Abstract
We report here the characterization of a bypass suppressor of pab1Delta which leads to a fourfold stabilization of the unstable MFA2 mRNA. Cloning of the wild-type gene for that suppressor reveals that it is identical to PAT1 (YCR077c), a gene whose product was reported to interact with Top2p. PAT1 is not an essential gene, but its deletion leads to a thermosensitive phenotype. Further analysis has shown that PAT1 is allelic with mrt1-3, a mutation previously reported to affect decapping and to bypass suppress pab1Delta, as is also the case for dcp1, spb8, and mrt3. Coimmunoprecipitation experiments show that Pat1p is associated with Spb8p. On sucrose gradients, the two proteins cosediment with fractions containing the polysomes. In the absence of Pat1p, however, Spb8p no longer cofractionates with the polysomes, while the removal of Spb8p leads to a sharp decrease in the level of Pat1p. Our results suggest that some of the factors involved in mRNA degradation could be associated with the mRNA that is still being translated, awaiting a specific signal to commit the mRNA to the degradation pathway.
Figures
FIG. 1
Characterization of the spb10 mutation and cloning of the PAT1 gene. (A) Complementation testing for thermosensitivity of the spb10-1 allele with various plasmids. Yeast cultures were spotted as serial 10-fold dilution onto yeast-peptone-dextrose (YPD) plates and grown for 3 days either at the permissive (25°C, right panel) or at the restrictive (37°C, left panel) temperature. First row, spb10-1 mutant strain; second row, the same strain transformed with the pFL44-A1 plasmid that contains the PAT1 gene; third row, the spb10-1 strain transformed with the pFL44-A1K plasmid that contains the mutated PAT1 gene. (B) Deletion of PAT1 leads to thermosensitivity and to an spb phenotype. A diploid strain, pat1Δ::TRP1/PAT1 pab1Δ::HIS3/PAB1 (pAS77 [p_PAB1 URA3 CEN_]), was constructed by genetic crossing and sporulated, and tetrads were dissected and analyzed. His+ spores were selected, and their growth was tested under different conditions as indicated.
FIG. 2
Sequence comparison between Pat1p from budding and fission yeasts. The sequence of Pat1p from S. cerevisiae (Sc) and a translated ORF from S. pombe (Sp) (accession number CAA17064) were aligned (Clustal W [46]). Five regions of higher similarity were chosen and named motifs I through V (shown in Roman numerals on the right). Residues that are either identical or chemically equivalent are boxed. Numbers at the top refer to the Pat1p sequence from S. cerevisiae. The dash represents a gap inserted in motif I to optimize the alignment.
FIG. 3
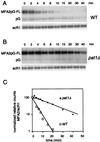
MFA2pG mRNA is stabilized in the pat1Δ strain. (A and B) Degradation of MFA2pG mRNA in wild-type (WT) (A) and pat1Δ (B) strains. Cells carrying the GAL::MFA2pG reporter gene were shifted from galactose- to glucose-containing medium to repress transcription. RNA was recovered from samples taken at the indicated times after repression and analyzed by Northern blotting. The top panel shows hybridization with an oligonucleotide (oRP140) recognizing the full-length MFA2pG mRNA and the pG degradation intermediate. The bottom panel shows the hybridization signal with the scR1 probe, which is used here as an internal control. (C) The level of full-length MFA2pG mRNA for each time point was quantitated by PhosphorImager (Molecular Dynamics) analysis, normalized to scR1, and then plotted on a log scale as a function of time.
FIG. 4
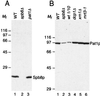
PAT1 is identical to MRT1. (A) A plasmid containing the wild-type (WT) PAT1 gene can support the growth of an mrt1-3 strain at 37°C. Tenfold serial dilutions of the indicated strains were spotted on yeast minimal medium plates lacking uracil and incubated at either 25 or 37°C as shown at the bottom. WT, a W303 derivative containing plasmid pRP485; +pFL44, yRP1066 (18) transformed with an empty pFL44 vector; +pFL44-A1, the same strain transformed with the plasmid containing the PAT1 gene. (B) Pat1p is not detected in an mrt1-3 haploid strain or in a diploid pat1Δ mrt1-3 double-mutant strain. Protein extracts (50 μg) were fractionated by SDS-PAGE (10% polyacrylamide), electroblotted onto nitrocellulose, and probed with antibodies raised against Pat1p. Lanes: 1 to 3, haploid strains; 4 to 8, diploid strains. Relevant genotypes are indicated above each lane. _M_r, molecular weight markers in thousands.
FIG. 5
The absence of Spb8p leads to a sharp decrease in the level of Pat1p. (A) Protein extracts (50 μg) prepared from the indicated strains were tested by Western blotting with the rabbit anti-Spb8p antibody. (B) Detection of Pat1p in various mutant strains. Protein extract (50 μg) prepared from each strain (5 μg for lane 3) was subjected to SDS-PAGE (10% polyacrylamide). The filters were probed with the anti-Pat1p antibody. Relevant genotypes are indicated above each lane. _M_r, molecular weight markers in thousands. WT, wild type.
FIG. 6
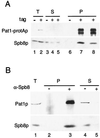
Spb8p and Pat1p are coimmunoprecipitated. (A) Immunoprecipitation of Pat1-protAp. Native extracts prepared either from a wild-type strain or from a PAT1-protA tagged strain were incubated with immunoglobulin G-Sepharose beads. After extensive washes, proteins were eluted from the beads and separated by SDS-PAGE (10% polyacrylamide). Western blotting was performed either with rabbit immunoglobulin G coupled to horseradish peroxidase to detect the protA-tagged protein (upper panel) or with an anti-Spb8p antibody (lower panel). Lanes: 1, 3, and 6, wild-type strain (tag −); 2, 4, 5, 7, and 8, PAT1-protA strain (tag +). Lanes 1 to 5 contain 1/40 the amount of protein extract used in the immunoprecipitation experiment. T, total extract; S, supernatant; P, pellet. Lanes 5 and 8 contain the supernatant and pellet, respectively, of the immunoprecipitation, washed with a buffer containing 0.5 M NaCl. (B) Immunoprecipitation of Spb8p. Native extracts prepared from strain yRP1070 transformed with pRP801 (see Materials and Methods) were treated with a preimmune serum and incubated with protein A-Sepharose to remove any nonspecific complexes. Then the supernatants were treated with the anti-Spb8p antibody or left untreated; this was followed by precipitation with protein A-Sepharose. Proteins from the pellets and the supernatants were analyzed by Western blotting with either the anti-Pat1p (upper panel) or the anti-Spb8p (lower panel) antibodies. Lane 1 contains 1/40 the amount of protein used in the immunoprecipitation (T); lanes 2 and 3 contain pellets of the immunoprecipitation (P); lanes 4 and 5 contain 1/40 of the supernatants (S). Lanes 2 and 4, control immunoprecipitation performed without serum (−); lanes 3 and 5, immunoprecipitation done in the presence of the anti-Spb8p antibodies (+).
FIG. 7
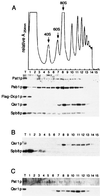
Spb8p and Pat1p cosediment with polysomes on sucrose gradients. (A) A total-cell extract prepared from strain yRP1070 transformed with pRP801, which expresses a tagged version of Dcp1p, was fractionated on a 7 to 50% linear sucrose gradient. (Top) A continuous record of the absorbance at 254 nm (A254nm) for the gradient is presented, with the top of the gradient on the left. The arrows indicate the peaks for the 40S, 60S, and 80S subunits. (Bottom) Western blot analysis of the 15 fractions collected from the sucrose gradient was performed with antibodies raised against the indicated proteins. T, total-cell extract. (B) A total-cell extract was prepared from a strain with PAT1 deleted and fractionated on a sucrose gradient as described for panel A. The 15 fractions were analyzed by Western blotting with the anti-Qsr1p (upper row) or the anti-Spb8p (bottom row) antibody. (C) As in panel B, except that extracts were prepared from a strain with SPB8 deleted and tested with the anti-Pat1p (upper row) or the anti-Qsr1p (bottom row) antibody.
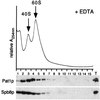
FIG. 8
Addition of EDTA results in a shift of Spb8p and Pat1p to the upper part of the sucrose gradient. Extracts were prepared and analyzed as described in the legend to Fig. 7, except that 40 mM EDTA was added to the breaking buffer and to the sucrose gradient solutions. (Top) Relative profile of the absorbance at 254 nm (A254nm) with the top of the gradient on the left. Arrows indicate the positions of the 40S and 60S subunits. (Bottom) Western blot analysis of the fractions using either the anti-Pat1p or the anti-Spb8p antibodies.
Similar articles
- Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast.
Wyers F, Minet M, Dufour ME, Vo LT, Lacroute F. Wyers F, et al. Mol Cell Biol. 2000 May;20(10):3538-49. doi: 10.1128/MCB.20.10.3538-3549.2000. Mol Cell Biol. 2000. PMID: 10779343 Free PMC article. - Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation.
Mangus DA, Amrani N, Jacobson A. Mangus DA, et al. Mol Cell Biol. 1998 Dec;18(12):7383-96. doi: 10.1128/MCB.18.12.7383. Mol Cell Biol. 1998. PMID: 9819425 Free PMC article. - The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3' termini from partial degradation.
He W, Parker R. He W, et al. Genetics. 2001 Aug;158(4):1445-55. doi: 10.1093/genetics/158.4.1445. Genetics. 2001. PMID: 11514438 Free PMC article. - Pat1 proteins: a life in translation, translation repression and mRNA decay.
Marnef A, Standart N. Marnef A, et al. Biochem Soc Trans. 2010 Dec;38(6):1602-7. doi: 10.1042/BST0381602. Biochem Soc Trans. 2010. PMID: 21118134 Review. - mRNA decapping activities and their biological roles.
LaGrandeur TE, Parker R. LaGrandeur TE, et al. Biochimie. 1996;78(11-12):1049-55. doi: 10.1016/s0300-9084(97)86729-6. Biochimie. 1996. PMID: 9150884 Review.
Cited by
- Expression profile and prognostic values of LSM family in skin cutaneous melanoma.
Sun X, Zhang J, Xiao C, Ge Z. Sun X, et al. BMC Med Genomics. 2022 Nov 12;15(1):238. doi: 10.1186/s12920-022-01395-6. BMC Med Genomics. 2022. PMID: 36371223 Free PMC article. - GC content shapes mRNA storage and decay in human cells.
Courel M, Clément Y, Bossevain C, Foretek D, Vidal Cruchez O, Yi Z, Bénard M, Benassy MN, Kress M, Vindry C, Ernoult-Lange M, Antoniewski C, Morillon A, Brest P, Hubstenberger A, Roest Crollius H, Standart N, Weil D. Courel M, et al. Elife. 2019 Dec 19;8:e49708. doi: 10.7554/eLife.49708. Elife. 2019. PMID: 31855182 Free PMC article. - Identification of novel mutations in ACT1 and SLA2 that suppress the actin-cable-overproducing phenotype caused by overexpression of a dominant active form of Bni1p in Saccharomyces cerevisiae.
Yoshiuchi S, Yamamoto T, Sakane H, Kadota J, Mochida J, Asaka M, Tanaka K. Yoshiuchi S, et al. Genetics. 2006 Jun;173(2):527-39. doi: 10.1534/genetics.105.055210. Epub 2006 Mar 17. Genetics. 2006. PMID: 16547104 Free PMC article. - Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication.
Mas A, Alves-Rodrigues I, Noueiry A, Ahlquist P, Díez J. Mas A, et al. J Virol. 2006 Jan;80(1):246-51. doi: 10.1128/JVI.80.1.246-251.2006. J Virol. 2006. PMID: 16352549 Free PMC article. - A role for Lsmlp in response to ultraviolet-radiation damage in Saccharomyces cerevisiae.
Spicakova T, McCann K, Brown JM. Spicakova T, et al. Radiat Res. 2008 Oct;170(4):411-21. doi: 10.1667/rr1477.1. Radiat Res. 2008. PMID: 19024647 Free PMC article.
References
- Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. - PubMed
- Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous