Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis - PubMed (original) (raw)
Comparative Study
Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis
K Oegema et al. J Cell Biol. 2000.
Abstract
We have characterized a human homologue of anillin, a Drosophila actin binding protein. Like Drosophila anillin, the human protein localizes to the nucleus during interphase, the cortex following nuclear envelope breakdown, and the cleavage furrow during cytokinesis. Anillin also localizes to ectopic cleavage furrows generated between two spindles in fused PtK(1) cells. Microinjection of antianillin antibodies slows cleavage, leading to furrow regression and the generation of multinucleate cells. GFP fusions that contain the COOH-terminal 197 amino acids of anillin, which includes a pleckstrin homology (PH) domain, form ectopic cortical foci during interphase. The septin Hcdc10 localizes to these ectopic foci, whereas myosin II and actin do not, suggesting that anillin interacts with the septins at the cortex. Robust cleavage furrow localization requires both this COOH-terminal domain and additional NH(2)-terminal sequences corresponding to an actin binding domain defined by in vitro cosedimentation assays. Endogenous anillin and Hcdc10 colocalize to punctate foci associated with actin cables throughout mitosis and the accumulation of both proteins at the cell equator requires filamentous actin. These results indicate that anillin is a conserved cleavage furrow component important for cytokinesis. Interactions with at least two other furrow proteins, actin and the septins, likely contribute to anillin function.
Figures
Figure 1
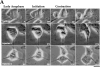
Characterization of an anillin homologue. (A) An alignment of the conserved COOH-terminal region of anillin homologues found in blast searches of the databases. The aligned sequences are human anillin (this report; GenBank accession No. AF273437); Drosophila anillin (Field and Alberts 1995; GenBank accession No. X89858); the products of the C. elegans genes K10B2.5 (GenBank accession No. T16604), Y43F8C.14 (GenBank accession No. T26874), and Y49E10.19 (GenBank accession No. T27053); and the product of the Drosophila gene CG4530 (GenBank accession No. AAF47044). The sequences were aligned with ClustalX (Thompson et al. 1997) using the default settings. The asterisk indicate positions where the amino acid is identical in all six sequences; a single dot indicates positions where the amino acids of all sequences fall into a weak similarity group; and a double dot indicates positions where the amino acids of all sequences fall into a strong similarity group. The PH domain is underlined. (B) A schematic comparison of Drosophila and human anillin homologues. The overall level of identity between the two proteins is 25%. The COOH-terminal third of the protein is the most conserved (∼36% identity). This region includes a PH domain (human, amino acids 985–1,110; Drosophila, amino acids 1,069–1,194), which is 54% identical between the two proteins. The minimal sequences required for actin binding (amino acids 258–340, indicated here in black) and bundling (amino acids 246–371) have been mapped for the Drosophila protein (Field and Alberts 1995). A larger region of the Drosophila protein (amino acids 127–371, shown here in medium gray) was found to stabilize actin binding and bundling. The locations of potential nuclear localization sequences, identified using the program PSORT II (Nakai and Horton 1999), are indicated with black triangles (human, amino acids 64, 66, 195, 786, 896, and 1,021; Drosophila, amino acids 996 and 1,105). Also conserved is a potential SH3 binding motif. There is one SH3 binding consensus in the human and two in the Drosophila sequence (indicated here with asterisks). (C) An affinity-purified antibody against hsanillin recognizes a single band in extracts of mammalian cells. A Western blot of cell extracts probed with affinity-purified antibodies to hsanillin. Extracts of BHK-21, BS-C-1, and HeLa cells were loaded. In all cases, a single band running at ∼180 kD is detected. (D) hsanillin, like the Drosophila protein, has a dynamic cell cycle–dependent localization pattern. Methanol-fixed BHK cells were stained for hsanillin. Three-dimensional widefield data was collected and deconvolved. The interphase cell is a projection of the entire stack; the other images are selected sections. In interphase cells, hsanillin is found primarily in the nucleus, upon nuclear envelope breakdown, hsanillin becomes enriched at the cortex. During cytokinesis, the protein localizes to the cleavage furrow. In late telophase, anillin is concentrated in the cortex surrounding the midbody.
Figure 2
Cytokinesis fails in cells microinjected with antianillin antibody. (A) Selected images from three time-lapse movies. Time (in seconds) from anaphase onset is shown in the bottom right-hand corner of each image. The top row shows an uninjected control cell. In the control, cytokinesis is almost complete in 10 min, and, within 13 min (776 s), the daughter cells have moved apart. The bottom two rows show two examples of cells injected with antianillin antibody. In both cases, furrowing initiates at approximately the same time as in the control cell; however, in each case, ingression stops and cell division fails. (B) Plots of the cell width near the equator with respect to time. Open triangles are control cells and filled circles are antibody-injected cells. The two cells shown in Fig. 2 A are plotted. Injected 1 contracts slowly, stops, and cytokinesis fails. Injected 2 constricts at a faster rate, but the furrow eventually relaxes, opens up, and cell division also fails. (C) Bar graph showing the difference in the maximum rate of contraction between control cells (0.035 ± 0.010 μm/s, n = 5) and cells injected with antianillin antibody (0.016 ± 0.0047 μm/s, n = 8). Error bars are the SD. Bar, 15 μm.
Figure 2
Cytokinesis fails in cells microinjected with antianillin antibody. (A) Selected images from three time-lapse movies. Time (in seconds) from anaphase onset is shown in the bottom right-hand corner of each image. The top row shows an uninjected control cell. In the control, cytokinesis is almost complete in 10 min, and, within 13 min (776 s), the daughter cells have moved apart. The bottom two rows show two examples of cells injected with antianillin antibody. In both cases, furrowing initiates at approximately the same time as in the control cell; however, in each case, ingression stops and cell division fails. (B) Plots of the cell width near the equator with respect to time. Open triangles are control cells and filled circles are antibody-injected cells. The two cells shown in Fig. 2 A are plotted. Injected 1 contracts slowly, stops, and cytokinesis fails. Injected 2 constricts at a faster rate, but the furrow eventually relaxes, opens up, and cell division also fails. (C) Bar graph showing the difference in the maximum rate of contraction between control cells (0.035 ± 0.010 μm/s, n = 5) and cells injected with antianillin antibody (0.016 ± 0.0047 μm/s, n = 8). Error bars are the SD. Bar, 15 μm.
Figure 3
Anillin is found in ectopic furrows that form between asters not previously connected by a spindle in fused PtK1 cells. (A–D) Selected images from a time-lapse sequence of a PtK1 heterokaryon, which forms an ectopic midbody between two independent mitotic spindles. Shortly after the onset of cytokinesis at each spindle equator (B, black arrows), an ectopic furrow formed (C, black arrowhead) and progressed to form a midbody (D). (E and F) Indirect immunofluorescence of the same cell fixed ∼5 min after image D. (E) Microtubule staining demonstrates that the ectopic midbody contains microtubules (white arrowhead), although fewer than the midbodies, associated with the spindle midzones (white arrows). (F) An analysis of anillin distribution reveals that it is always present at all midbodies including the ectopic (white arrowheads). (G) Merged images of microtubules (green), anillin (red), and DNA (blue). Bars, 10 μm.
Figure 4
Hsanillin domain analysis. (A) Schematic of the regions of human anillin that were fused to GFP for transfection into BHK cells. All fusions shown here and in the images in Fig. 5 had GFP fused to their NH2 terminus. However, the full-length protein as well as the regions 455–724, 731–1,125, and 983–1,125 were also tested with GFP fused to their COOH terminus. In all cases, these constructs gave the same localization as the NH2-terminal fusions. The chart on the right summarizes the localization of the tested fragments. For examples, see Fig. 5. (B) Western blot of cells transfected with each of the NH2-terminal GFP fusions. Samples were fractionated on a 12% SDS-PAGE gel, transferred to nitrocellulose, and probed with an antibody to GFP.
Figure 5
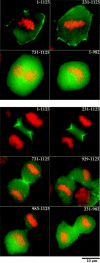
Both the actin binding domain and the PH domain are required for recruitment of anillin to the cortex and for efficient recruitment to the cleavage furrow during cytokinesis. BHK cells were transfected with the indicated GFP constructs and were fixed and stained for DNA. (Top) Cortical recruitment of selected fragments of anillin fused to GFP during metaphase. Full-length anillin (1–1,125) and a fragment, which deletes the first 231 amino acids, are efficiently recruited to the cortex during metaphase. No cortical enrichment of GFP fusions, which delete the actin binding region (731–1,125) or the PH domain (1–982), is observed. (Bottom) Full-length anillin (1–1,125) and a fragment that deletes the NH2 terminus (231–1,125) are efficiently recruited to the cleavage furrow during cytokinesis. The COOH terminus (731–1,125) is also recruited to the cleavage furrow, but less efficiently. The PH domain alone (983–1,125) or a slightly longer region that contains the PH domain (the extended PH domain, 929–1,125) are not sufficient to direct GFP to the cleavage furrow. GFP fusions that delete only the PH domain (1–982; not shown) or the PH domain and the first 230 amino acids (231–982) were very weakly enriched in the cleavage furrow.
Figure 6
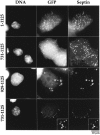
GFP fusions with regions of anillin that contain the COOH-terminal 197 amino acids form ectopic cortical foci during interphase. The septin Hcdc10 also localizes to these foci. BHK cells were transfected with the indicated GFP constructs, fixed, and stained for DNA and the septin Hcdc10. Fusions of GFP with full-length anillin (1–1,125) or with the anillin COOH terminus (731–1,125) form ectopic cortical foci when overexpressed in interphase cells. The septin Hcdc10 localizes to these foci. A fusion of GFP with the COOH-terminal 197 amino acids of anillin (929–1,125) is sufficient to form ectopic foci that colocalize Hcdc10, but these foci tend to be a bit larger and often appear to be beneath the cortex. (Bottom row) Fusions of GFP with the COOH-terminal 395 amino acids of anillin (731–1,125) also often formed ringlike foci. Hcdc10 also colocalized to these rings.
Figure 7
Anillin and Hcdc10 are found in punctate foci that localize along actin cables during mitosis in BHK cells. BHK cells were fixed and stained for actin, Hcdc10, anillin, and DNA. Three-dimensional widefield data sets were collected and deconvolved. Shown are projections of entire cells. During metaphase and early anaphase, actin is found in cables that often appear to encircle the cell. During late anaphase, as the cell initiates contraction, the actin cables appear to align parallel to the spindle axis. As contraction proceeds, more actin cables within the cleavage furrow are observed perpendicular to the spindle axis but, within the actively contracting region near the base of the cell, the actin cables are still parallel to the spindle axis. During all phases of mitosis, anillin and Hcdc10 colocalize to foci that align along actin cables.
Figure 8
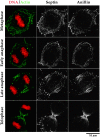
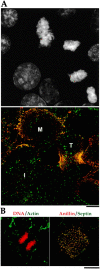
Anillin and the septin Hcdc10 colocalize on the cortex of latrunculin-treated cells but do not accumulate in the cleavage furrow. (A) A typical field of BHK cells fixed and stained for DNA (top), anillin (bottom, red), and Hcdc10 (bottom, green). Three-dimensional widefield data sets were collected and deconvolved. A projection of the field is shown. During interphase (I) Hcdc10 is present in punctate cortical structures on the cortex. Anillin is in the nuclei of most, but not all, interphase cells. Note that the nuclear staining is somewhat weaker in formaldehyde-fixed cells compared with the methanol-fixed cells shown in Fig. 1. At metaphase (M), anillin concentrates at the cortex where it colocalizes with septin foci. At telophase (T), foci containing anillin and Hcdc10 are concentrated in the cleavage furrow. (B) BHK cells grown on coverslips were treated with 2 μg/ml latrunculin A (a gift from Miranda Sanders and Phil Crews, University of California, Santa Cruz) or latrunculin B (Calbiochem) in normal culture media for 60 min before processing for immunofluorescence. In the left panel, a merge of DNA (red) and actin (green) is shown. In the right panels, a merge of anillin (red) and the septin Hcdc10 (green) is shown. Images were collected as described for Fig. 7 and are projections of entire cells. When the actin is depolymerized by latrunculin treatment, neither Hcdc10 nor anillin are recruited to the cell equator during mitosis; instead, they colocalize to large cortical spots.
Similar articles
- Stabilization of the actomyosin ring enables spermatocyte cytokinesis in Drosophila.
Goldbach P, Wong R, Beise N, Sarpal R, Trimble WS, Brill JA. Goldbach P, et al. Mol Biol Cell. 2010 May 1;21(9):1482-93. doi: 10.1091/mbc.e09-08-0714. Epub 2010 Mar 17. Mol Biol Cell. 2010. PMID: 20237160 Free PMC article. - Cleavage furrow organization requires PIP(2)-mediated recruitment of anillin.
Liu J, Fairn GD, Ceccarelli DF, Sicheri F, Wilde A. Liu J, et al. Curr Biol. 2012 Jan 10;22(1):64-9. doi: 10.1016/j.cub.2011.11.040. Epub 2011 Dec 22. Curr Biol. 2012. PMID: 22197245 - Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis.
Piekny AJ, Glotzer M. Piekny AJ, et al. Curr Biol. 2008 Jan 8;18(1):30-6. doi: 10.1016/j.cub.2007.11.068. Epub 2007 Dec 27. Curr Biol. 2008. PMID: 18158243 - Anillin: a pivotal organizer of the cytokinetic machinery.
Hickson GR, O'Farrell PH. Hickson GR, et al. Biochem Soc Trans. 2008 Jun;36(Pt 3):439-41. doi: 10.1042/BST0360439. Biochem Soc Trans. 2008. PMID: 18481976 Free PMC article. Review. - How to scaffold the contractile ring for a safe cytokinesis - lessons from Anillin-related proteins.
D'Avino PP. D'Avino PP. J Cell Sci. 2009 Apr 15;122(Pt 8):1071-9. doi: 10.1242/jcs.034785. J Cell Sci. 2009. PMID: 19339546 Review.
Cited by
- Anillin localization suggests distinct mechanisms of division plane specification in mouse oogenic meiosis I and II.
Sharif B, Fadero T, Maddox AS. Sharif B, et al. Gene Expr Patterns. 2015 Mar;17(2):98-106. doi: 10.1016/j.gep.2015.03.002. Epub 2015 Mar 26. Gene Expr Patterns. 2015. PMID: 25818309 Free PMC article. - Specification of Architecture and Function of Actin Structures by Actin Nucleation Factors.
Skau CT, Waterman CM. Skau CT, et al. Annu Rev Biophys. 2015;44:285-310. doi: 10.1146/annurev-biophys-060414-034308. Annu Rev Biophys. 2015. PMID: 26098516 Free PMC article. Review. - Dysregulated ANLN reveals immune cell landscape and promotes carcinogenesis by regulating the PI3K/Akt/mTOR pathway in clear cell renal cell carcinoma.
Gao M, Tuo Z, Jiang Z, Chen Z, Wang J. Gao M, et al. Heliyon. 2023 Dec 9;10(1):e23522. doi: 10.1016/j.heliyon.2023.e23522. eCollection 2024 Jan 15. Heliyon. 2023. PMID: 38173514 Free PMC article. - Septin/anillin filaments scaffold central nervous system myelin to accelerate nerve conduction.
Patzig J, Erwig MS, Tenzer S, Kusch K, Dibaj P, Möbius W, Goebbels S, Schaeren-Wiemers N, Nave KA, Werner HB. Patzig J, et al. Elife. 2016 Aug 9;5:e17119. doi: 10.7554/eLife.17119. Elife. 2016. PMID: 27504968 Free PMC article. - ANLN-induced EZH2 upregulation promotes pancreatic cancer progression by mediating miR-218-5p/LASP1 signaling axis.
Wang A, Dai H, Gong Y, Zhang C, Shu J, Luo Y, Jiang Y, Liu W, Bie P. Wang A, et al. J Exp Clin Cancer Res. 2019 Aug 8;38(1):347. doi: 10.1186/s13046-019-1340-7. J Exp Clin Cancer Res. 2019. PMID: 31395079 Free PMC article.
References
- Beites C.L., Xie H., Bowser R., Trimble W.S. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 1999;2:434–439. - PubMed
- Benzanilla M., Wilson J.M., Pollard T.D. Fission yeast myosin-II isoforms assemble into contractile rings at distinct times during mitosis. Curr. Biol. 2000;10:397–400. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous