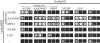The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein - PubMed (original) (raw)
Comparative Study
The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein
V P Efimov et al. J Cell Biol. 2000.
Abstract
The nudF gene of the filamentous fungus Aspergillus nidulans acts in the cytoplasmic dynein/dynactin pathway and is required for distribution of nuclei. NUDF protein, the product of the nudF gene, displays 42% sequence identity with the human protein LIS1 required for neuronal migration. Haploinsufficiency of the LIS1 gene causes a malformation of the human brain known as lissencephaly. We screened for multicopy suppressors of a mutation in the nudF gene. The product of the nudE gene isolated in the screen, NUDE, is a homologue of the nuclear distribution protein RO11 of Neurospora crassa. The highly conserved NH(2)-terminal coiled-coil domain of the NUDE protein suffices for protein function when overexpressed. A similar coiled-coil domain is present in several putative human proteins and in the mitotic phosphoprotein 43 (MP43) of X. laevis. NUDF protein interacts with the Aspergillus NUDE coiled-coil in a yeast two-hybrid system, while human LIS1 interacts with the human homologue of the NUDE/RO11 coiled-coil and also the Xenopus MP43 coiled-coil. In addition, NUDF coprecipitates with an epitope-tagged NUDE. The fact that NUDF and LIS1 interact with the same protein domain strengthens the notion that these two proteins are functionally related.
Figures
Figure 1
(A) Suppression of nud mutants by extra copies of the nudE and nudF6 genes. The indicated mutants (all are conditional, temperature sensitive) were transformed with either the empty vector pAid or pAid clones bearing nudE and nudF6 genes (plasmids recovered in the multicopy suppressor screen) and grown at 32°C. Transformants were gridded on YAG plates with or without KCl and incubated at 43°C or 32°C for 2 d. All strains have different color of conidia: yellow for nudF7, green (wild-type) for nudF6, chartreuse for nudA1, and white for nudC3. The intensity of the colony color is proportional to the number of conidia produced. Four independent transformants of the nudF7 mutant are shown for each plasmid to demonstrate reproducibility of phenotypes. (B) Complementation of the nudE deletion and the nudF7 mutant by extra copies of nudE, the nudE NH2-terminal domain, and nudE chimeras carrying coiled-coil regions from human and frog proteins, respectively. Strains were transformed with nudE variants in pAid vector and grown at 43°C. Numbers refer to amino acid residues of NUDE protein expressed by the constructs (see Fig. 3 for detailed amino acid sequences). Due to the presence of a fawn color marker, conidiating and unconidiating ΔnudE colonies have very similar colors at this temperature.
Figure 1
(A) Suppression of nud mutants by extra copies of the nudE and nudF6 genes. The indicated mutants (all are conditional, temperature sensitive) were transformed with either the empty vector pAid or pAid clones bearing nudE and nudF6 genes (plasmids recovered in the multicopy suppressor screen) and grown at 32°C. Transformants were gridded on YAG plates with or without KCl and incubated at 43°C or 32°C for 2 d. All strains have different color of conidia: yellow for nudF7, green (wild-type) for nudF6, chartreuse for nudA1, and white for nudC3. The intensity of the colony color is proportional to the number of conidia produced. Four independent transformants of the nudF7 mutant are shown for each plasmid to demonstrate reproducibility of phenotypes. (B) Complementation of the nudE deletion and the nudF7 mutant by extra copies of nudE, the nudE NH2-terminal domain, and nudE chimeras carrying coiled-coil regions from human and frog proteins, respectively. Strains were transformed with nudE variants in pAid vector and grown at 43°C. Numbers refer to amino acid residues of NUDE protein expressed by the constructs (see Fig. 3 for detailed amino acid sequences). Due to the presence of a fawn color marker, conidiating and unconidiating ΔnudE colonies have very similar colors at this temperature.
Figure 3
(A) Homology between A. nidulans NUDE protein and N. crassa RO11 protein. Shaded residues are predicted to form a coiled-coil structure. The probabilities of coiled-coil formation are 0.9–1 for the NH2-terminal region and 0.7–0.8 for the central region. Probabilities of coiled-coil formation were calculated using program COILS version 2.1 (MTIDK matrix, unweighted a and d positions, window = 28) at http://www.ch. embnet.org/software/COILS_form.html (Lupas et al. 1991). Assigning 2.5-fold weights to a and d positions slightly changes the probabilities at coiled-coil ends and increases probability for the central coiled-coil to 0.9–1. (B) Alignment between coiled-coil regions of an ORF predicted from human EST with GenBank accession number AA424443, X. laevis MP43 (accession number U95097), A. nidulans NUDE (accession number AF085679), and N. crassa RO11 (accession number AF015560). The underlined residues are predicted to form coiled-coil structure with probabilities >0.9. The regions between arrowheads were used in sequence exchange experiments to test if human or X. laevis coiled-coil regions can substitute for the NUDE coiled-coil. The percentages of identical (similar) residues for the underlined coiled-coil regions are 69% (82%) between NUDE and RO11, 38% (51%) between NUDE and AA424443, 39% (51%) between NUDE and MP43, 72% (83%) between AA424443 and MP43, 40% (52%) between RO11 and MP43, 40% (52%) between RO11 and AA424443.
Figure 3
(A) Homology between A. nidulans NUDE protein and N. crassa RO11 protein. Shaded residues are predicted to form a coiled-coil structure. The probabilities of coiled-coil formation are 0.9–1 for the NH2-terminal region and 0.7–0.8 for the central region. Probabilities of coiled-coil formation were calculated using program COILS version 2.1 (MTIDK matrix, unweighted a and d positions, window = 28) at http://www.ch. embnet.org/software/COILS_form.html (Lupas et al. 1991). Assigning 2.5-fold weights to a and d positions slightly changes the probabilities at coiled-coil ends and increases probability for the central coiled-coil to 0.9–1. (B) Alignment between coiled-coil regions of an ORF predicted from human EST with GenBank accession number AA424443, X. laevis MP43 (accession number U95097), A. nidulans NUDE (accession number AF085679), and N. crassa RO11 (accession number AF015560). The underlined residues are predicted to form coiled-coil structure with probabilities >0.9. The regions between arrowheads were used in sequence exchange experiments to test if human or X. laevis coiled-coil regions can substitute for the NUDE coiled-coil. The percentages of identical (similar) residues for the underlined coiled-coil regions are 69% (82%) between NUDE and RO11, 38% (51%) between NUDE and AA424443, 39% (51%) between NUDE and MP43, 72% (83%) between AA424443 and MP43, 40% (52%) between RO11 and MP43, 40% (52%) between RO11 and AA424443.
Figure 2
Extra copies of the nudE gene do not change the level of NUDF protein in the nudF7 mutant. Total protein extracts were prepared from nudF7 and nudA1 mutants carrying indicated plasmids and NUDF protein levels were analyzed by Western blotting with the anti-NUDF antibody. Strains were grown at 43°C in YG medium. A nudA1 mutant was included as a control of the wild-type level of NUDF. The amount of NUDF protein in the nudF6 mutant is less than in nudF7 (Xiang et al. 1995a).
Figure 4
NUDE and NUDF proteins interact in a two-hybrid system. S. cerevisiae strain AH109 was transformed with pairwise combinations of plasmids expressing indicated proteins as fusions with either the Gal4p DNA binding (Gal4pDBD) or Gal4p activating (Gal4pAD) domains. NUDE, AA424443 and MP43 refer to the NH2-terminal domains of A. nidulans NUDE protein (residues 1–195) and corresponding fusions with human and X. laevis homologues, respectively (Fig. 3 B). For each pair of plasmids, growth on three media is shown (from left to right): SD/-Leu/-Trp/-His; SD/-Leu/-Trp/-His with 3 mM 3-AT; SD/-Leu/-Trp/-Ade. The first two media select for the expression of the HIS3 reporter gene, while the third medium selects for the expression of the ADE2 reporter gene. Growth in the absence of histidine or adenine is expected to result from interactions between proteins encoded by plasmids. Note that Gal4pDBD fusions with the coiled-coil domains activate the HIS3 gene expression in the absence of any interactions. 3 mM 3-AT (3-amino-1,2,4,-triazole), a competitive inhibitor of the HIS3 gene product, suppresses the resulting background growth.
Figure 5
NUDE protein tagged with VSV-G epitopes coprecipitates with NUDF in A. nidulans protein extracts. (A) Total protein extracts from four independently isolated NUDE::(VSV-G)6 strains (#8, 12, 15, 17) and the parent strain with the wild-type nudE gene were analyzed by SDS-PAGE (4–20%) and immunoblotting with P5D4 antibody (left). Ponceau S staining of the membrane after protein transfer is shown on the right. (B) Same samples as in A (tagged strain #17 and untagged control) after better separation on a 10% SDS-PAGE. (C) The left panel shows immunoblotting of proteins precipitated by P5D4 antibodies coupled to Sepharose beads from nudE::(VSV-G)6 and nudE + extracts with P5D4 and anti-NUDF antibodies. The right panel shows silver staining of the same samples.
Similar articles
- The LIS1-related protein NUDF of Aspergillus nidulans and its interaction partner NUDE bind directly to specific subunits of dynein and dynactin and to alpha- and gamma-tubulin.
Hoffmann B, Zuo W, Liu A, Morris NR. Hoffmann B, et al. J Biol Chem. 2001 Oct 19;276(42):38877-84. doi: 10.1074/jbc.M106610200. Epub 2001 Aug 16. J Biol Chem. 2001. PMID: 11509576 Retracted. - Nudf, a fungal homolog of the human LIS1 protein, functions as a dimer in vivo.
Ahn C, Morris NR. Ahn C, et al. J Biol Chem. 2001 Mar 30;276(13):9903-9. doi: 10.1074/jbc.M010233200. Epub 2000 Dec 29. J Biol Chem. 2001. PMID: 11134054 - CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans.
Efimov VP, Zhang J, Xiang X. Efimov VP, et al. Mol Biol Cell. 2006 Apr;17(4):2021-34. doi: 10.1091/mbc.e05-11-1084. Epub 2006 Feb 8. Mol Biol Cell. 2006. PMID: 16467375 Free PMC article. - Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development.
Wynshaw-Boris A. Wynshaw-Boris A. Clin Genet. 2007 Oct;72(4):296-304. doi: 10.1111/j.1399-0004.2007.00888.x. Clin Genet. 2007. PMID: 17850624 Review. - Nuclear migration, nucleokinesis and lissencephaly.
Morris NR, Efimov VP, Xiang X. Morris NR, et al. Trends Cell Biol. 1998 Dec;8(12):467-70. doi: 10.1016/s0962-8924(98)01389-0. Trends Cell Biol. 1998. PMID: 9861667 Review.
Cited by
- Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent.
Zhang J, Li S, Fischer R, Xiang X. Zhang J, et al. Mol Biol Cell. 2003 Apr;14(4):1479-88. doi: 10.1091/mbc.e02-08-0516. Mol Biol Cell. 2003. PMID: 12686603 Free PMC article. - Identification of a novel dynein binding domain in nudel essential for spindle pole organization in Xenopus egg extract.
Wang S, Zheng Y. Wang S, et al. J Biol Chem. 2011 Jan 7;286(1):587-93. doi: 10.1074/jbc.M110.181578. Epub 2010 Nov 5. J Biol Chem. 2011. PMID: 21056974 Free PMC article. - A genome-scale RNAi screen for genetic interactors of the dynein co-factor nud-2 in Caenorhabditis elegans.
Rocha H, Maia AF, Gassmann R. Rocha H, et al. Sci Data. 2018 Mar 20;5:180047. doi: 10.1038/sdata.2018.47. Sci Data. 2018. PMID: 29557975 Free PMC article. - Ndel1 disfavors dynein-dynactin-adaptor complex formation in two distinct ways.
Garrott SR, Gillies JP, Siva A, Little SR, El Jbeily R, DeSantis ME. Garrott SR, et al. J Biol Chem. 2023 Jun;299(6):104735. doi: 10.1016/j.jbc.2023.104735. Epub 2023 Apr 21. J Biol Chem. 2023. PMID: 37086789 Free PMC article. - Shootin1 acts in concert with KIF20B to promote polarization of migrating neurons.
Sapir T, Levy T, Sakakibara A, Rabinkov A, Miyata T, Reiner O. Sapir T, et al. J Neurosci. 2013 Jul 17;33(29):11932-48. doi: 10.1523/JNEUROSCI.5425-12.2013. J Neurosci. 2013. PMID: 23864681 Free PMC article.
References
- Ahmad F.J., Hughey J., Wittmann T., Hyman A., Greaser M., Baas P.W. Motor proteins regulate force interactions between microtubules and microfilaments in the axon. Nat. Cell. Biol. 2000;2:276–280. - PubMed
- Aleksenko A., Clutterbuck A.J. Autonomous plasmid replication in Aspergillus nidulansAMA1 and MATE elements. Fungal. Genet. Biol. 1997;21:373–387. - PubMed
- Brunati A.M., James P., Guerra B., Ruzzene M., Donella-Deana A., Pinna L.A. The spleen protein-tyrosine kinase TPK-IIB is highly similar to the catalytic domain of p72syk. Eur. J. Biochem. 1996;240:400–407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous