Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells - PubMed (original) (raw)
Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells
K Narayanan et al. J Virol. 2000 Sep.
Abstract
Coronavirus contains three envelope proteins, M, E and S, and a nucleocapsid, which consists of genomic RNA and N protein, within the viral envelope. We studied the macromolecular interactions involved in coronavirus assembly in cells infected with a murine coronavirus, mouse hepatitis virus (MHV). Coimmunoprecipitation analyses demonstrated an interaction between N protein and M protein in infected cells. Pulse-labeling experiments showed that newly synthesized, unglycosylated M protein interacted with N protein in a pre-Golgi compartment, which is part of the MHV budding site. Coimmunoprecipitation analyses further revealed that M protein interacted with only genomic-length MHV mRNA, mRNA 1, while N protein interacted with all MHV mRNAs. These data indicated that M protein interacted with the nucleocapsid, consisting of N protein and mRNA 1, in infected cells. The M protein-nucleocapsid interaction occurred in the absence of S and E proteins. Intracellular M protein-N protein interaction was maintained after removal of viral RNAs by RNase treatment. However, the M protein-N protein interaction did not occur in cells coexpressing M protein and N protein alone. These data indicated that while the M protein-N protein interaction, which is independent of viral RNA, occurred in the M protein-nucleocapsid complex, some MHV function(s) was necessary for the initiation of M protein-nucleocapsid interaction. The M protein-nucleocapsid interaction, which occurred near or at the MHV budding site, most probably represented the process of specific packaging of the MHV genome into MHV particles.
Figures
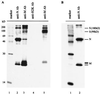
FIG. 1
Interaction between N protein and M protein in MHV-infected cells. (A) DBT cells were infected with MHV-JHM, and intracellular proteins were labeled with Tran[35S] label from 8 to 8.5 h p.i. (lanes 2 to 4) or with [3H]glucosamine from 6.5 to 8.5 h p.i. (lane 5). The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 2), an anti-M protein monoclonal antibody (lanes 3, 5) or an anti-H2K monoclonal antibody (lane 4), and viral proteins were analyzed by SDS–15% PAGE. Lane 1, 14C-labeled protein size marker. (B) MHV-JHM-infected DBT cells were pulse-labeled with Tran[35S] label for 5 min at 8 h p.i., and intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 2). Lane 1, 14C-labeled protein size marker. Ab, antibody.
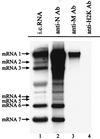
FIG. 2
Specific M protein-mRNA 1 interaction in MHV-infected cells. MHV-JHM-infected DBT cells were labeled with 32Pi from 6 to 8 h p.i. in the presence of actinomycin D, and cytoplasmic protein lysates were prepared. The intracellular (i.c.) proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 2), an anti-M protein monoclonal antibody (lane 3), or an anti-H2K monoclonal antibody (lane 4). MHV-specific RNAs were extracted from the immunoprecipitated samples and analyzed by agarose-formaldehyde gel electrophoresis. Lane 1, virus-specific RNAs extracted from 32P-labeled MHV-infected cells at 8 h p.i.
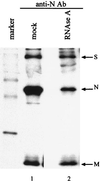
FIG. 3
Interaction between N protein and M protein after RNase A treatment. MHV-JHM-infected DBT cells were labeled with Tran[35S] label from 8 to 8.5 h p.i., and cytoplasmic lysates were prepared. Equal volumes of the lysates were either treated with RNase A (lane 2) or mock treated (lane 1) for 15 min at room temperature. The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody and analyzed by SDS–15% PAGE.
FIG. 4
Analysis of interaction between expressed N protein and M protein. (A) DBT cells that were infected with SinN pseudovirions (lanes 1 and 2) or SinM pseudovirions (lanes 3 and 4) were labeled with Tran[35S] label from 5 to 5.5 h p.i. The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lanes 1 and 4) or an anti-M protein monoclonal antibody (lanes 2 and 3), and viral proteins were analyzed by SDS–15% PAGE. (B) Equal volumes of 35S-labeled intracellular protein lysates from DBT cells, infected with SinM pseudovirions alone and SinN pseudovirions alone, were mixed, and intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 1), an anti-M protein monoclonal antibody (lane 2), or an anti-H2K monoclonal antibody (lane 3). The viral proteins were analyzed by SDS–12% PAGE. (C) DBT cells were coinfected with SinM pseudovirions and SinN pseudovirions, and intracellular proteins were labeled with Tran[35S] label from 5 to 5.5 h p.i. The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lane 1), an anti-M protein monoclonal antibody (lane 2), or an anti-H2K monoclonal antibody (lane 3). Viral proteins were analyzed by SDS–15% PAGE. The marked protein bands (indicated by arrows and an asterisk) are non-MHV proteins.
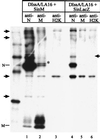
FIG. 5
Interaction between the nucleocapsid and M protein in the absence of S and E proteins. DBT cells were infected with DIssA/LA16 at 39.5°C. At 3.5 h post-DIssA/LA16 infection, cells were superinfected with either SinM pseudovirions (lanes 1 to 3) or SinLacZ pseudovirions (lanes 4 to 6) and incubated at 39.5°C. The intracellular proteins were labeled with Tran[35S] label from 8.5 to 9 h post-DIssA/LA16 infection, and cytoplasmic lysates were prepared. The intracellular proteins were immunoprecipitated with an anti-N protein monoclonal antibody (lanes 1 and 4), an anti-M protein monoclonal antibody (lanes 2 and 5), or an anti-H2K monoclonal antibody (lanes 3 and 6). The viral proteins were analyzed by SDS–12% PAGE. The 14C-labeled protein size marker is shown on the left of the gel. The marked protein bands (indicated by arrows and an asterisk) are non-MHV proteins.
FIG. 6
Specific interaction of M protein with DIssA RNA in the absence of S and E proteins. DBT cells were infected with DIssA/LA16 at 39.5°C. At 3.5 h post-DIssA/LA16 infection, cells were superinfected with either SinM pseudovirions (lanes 1 and 2) or SinLacZ pseudovirions (lanes 3 and 4) and incubated at 39.5°C. At 9 h post-DIssA/LA16 infection, cytoplasmic lysates were prepared and separated into two groups. Intracellular RNAs (lanes 1 and 3) were extracted from one group of lysates. An anti-M protein monoclonal antibody was added to another group, and immunoprecipitation was performed. RNAs were extracted from the immunoprecipitated samples (lanes 2 and 4). Extracted RNAs were separated by 1% agarose gel electrophoresis, and DIssA RNA was detected by Northern blot analysis. The part of the autoradiogram that contains DIssA RNA is indicated.
Similar articles
- Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus.
Kuo L, Masters PS. Kuo L, et al. J Virol. 2002 May;76(10):4987-99. doi: 10.1128/jvi.76.10.4987-4999.2002. J Virol. 2002. PMID: 11967315 Free PMC article. - Characterization of N protein self-association in coronavirus ribonucleoprotein complexes.
Narayanan K, Kim KH, Makino S. Narayanan K, et al. Virus Res. 2003 Dec;98(2):131-40. doi: 10.1016/j.virusres.2003.08.021. Virus Res. 2003. PMID: 14659560 Free PMC article. - Analyses of Coronavirus Assembly Interactions with Interspecies Membrane and Nucleocapsid Protein Chimeras.
Kuo L, Hurst-Hess KR, Koetzner CA, Masters PS. Kuo L, et al. J Virol. 2016 Apr 14;90(9):4357-4368. doi: 10.1128/JVI.03212-15. Print 2016 May. J Virol. 2016. PMID: 26889024 Free PMC article. - [Coronaviruses].
Taguchi F. Taguchi F. Uirusu. 2011 Dec;61(2):205-10. doi: 10.2222/jsv.61.205. Uirusu. 2011. PMID: 22916567 Review. Japanese. - Coronavirus genomic RNA packaging.
Masters PS. Masters PS. Virology. 2019 Nov;537:198-207. doi: 10.1016/j.virol.2019.08.031. Epub 2019 Aug 30. Virology. 2019. PMID: 31505321 Free PMC article. Review.
Cited by
- Porcine Epidemic Diarrhea Virus 3C-Like Protease-Mediated Nucleocapsid Processing: Possible Link to Viral Cell Culture Adaptability.
Jaru-Ampornpan P, Jengarn J, Wanitchang A, Jongkaewwattana A. Jaru-Ampornpan P, et al. J Virol. 2017 Jan 3;91(2):e01660-16. doi: 10.1128/JVI.01660-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27807240 Free PMC article. - Subcellular localization and membrane association of SARS-CoV 3a protein.
Yuan X, Li J, Shan Y, Yang Z, Zhao Z, Chen B, Yao Z, Dong B, Wang S, Chen J, Cong Y. Yuan X, et al. Virus Res. 2005 May;109(2):191-202. doi: 10.1016/j.virusres.2005.01.001. Virus Res. 2005. PMID: 15763150 Free PMC article. - Importance of the penultimate positive charge in mouse hepatitis coronavirus A59 membrane protein.
Verma S, Lopez LA, Bednar V, Hogue BG. Verma S, et al. J Virol. 2007 May;81(10):5339-48. doi: 10.1128/JVI.02427-06. Epub 2007 Feb 28. J Virol. 2007. PMID: 17329345 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources