The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages - PubMed (original) (raw)
The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages
K L Talks et al. Am J Pathol. 2000 Aug.
Abstract
The cellular response to hypoxia includes the hypoxia-inducible factor-1 (HIF-1)-induced transcription of genes involved in diverse processes such as glycolysis and angiogenesis. Induction of the HIF-regulated genes, as a consequence of the microenvironment or genetic changes, is known to have an important role in the growth of experimental tumors. Hypoxia-inducible factors 1alpha and 2alpha (HIF-1alpha and HIF-2alpha) are known to dimerize with the aryl hydrocarbon receptor nuclear translocator in mediating this response. Because regulation of the alpha chain protein level is a primary determinant of HIF activity, our aim was to investigate the distribution of HIF-1alpha and HIF-2alpha by immunohistochemistry in normal and pathological tissues using monoclonal antibodies (mAb). We raised a new mAb to detect HIF-1alpha, designated 122, and used our previously validated mAb 190b to HIF-2alpha. In the majority of solid tumors examined, including bladder, brain, breast, colon, ovarian, pancreatic, prostate, and renal carcinomas, nuclear expression of HIF-1alpha and -2alpha was observed in varying subsets of the tumor cells. HIF-2alpha was also strongly expressed by subsets of tumor-associated macrophages, sometimes in the absence of any tumor cell expression. Less frequently staining was observed in other stromal cells within the tumors and in normal tissue adjacent to tumor margins. In contrast, in normal tissue neither molecule was detectable except within subsets of bone marrow macrophages, where HIF-2alpha was strongly expressed.
Figures
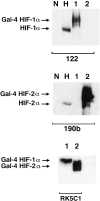
Figure 1.
Western blots showing specificity of mAbs used for immunostaining. Whole cell extracts were prepared from HeLa cells cultured in parallel for 4 hours in normoxia (N) and 0.1% hypoxia (H) and COS-1 cells transfected with either pGN/HIF-1α28–826 (lane 1) or pGN/HIF-2α19–870 (lane 2). Transfection resulted in the expression of fusion proteins between the N-terminal Gal4 DNA binding domain and the indicated amino acids of the respective HIF α chains. Preliminary analysis (not shown) using the Gal4 mAb established the amount of each COS extract required to give approximately equal Gal4 signal, indicating similar amounts of HIF-1α and HIF-2α fusion proteins. These were loaded with 50 μg of HeLa extracts and separated by SDS-PAGE, transferred to PVDF membrane and analyzed in parallel using mAb 122 (HIF-1α), 190b (HIF-2α), and RK5C1 (Gal4) as primary antibodies. mAb 122 detects a single band in extracts from COS cells transfected with pGN/HIF-1α28–826 and no bands in extracts from COS cells transfected with pGN/HIF-2α19–870, whereas the opposite pattern is seen with mAb 190b. Detection of comigrating bands with an antibody to the GAL4 DNA binding domain (RK5C1) confirms the identity of these bands as the respective fusion proteins. The antibodies to HIF-1α and HIF-2α recognize hypoxically inducible proteins of 135 kd and 110 kd, respectively, in the HeLa cell extracts, compatible with the known migration of endogenous HIF-1α and HIF-2α on SDS-PAGE.
Figure 2.
HIF-1α and HIF-2α protein detection by peroxidase immunohistochemistry on paraffin-embedded material. COS-1 cells were transfected with either pGN/HIF-1α28–826 (A and E) or pGN/HIF-2α19–870 (B and F) resulting in the expression of fusion proteins in a subset of the cells. Cells were then fixed in formalin and processed into paraffin-embedded blocks in a manner analogous to the handling of diagnostic pathological specimens. Likewise HT1080 cells were cultured in parallel for 4 hours in normoxia or 0.1% hypoxia and processed to produce a normoxic cell pellet (C and G) and a hypoxic cell pellet (D and H). Nuclear staining was observed with mAb 122 (A−D) only in the HIF-1α transfectants (A) and the hypoxic cell pellet (D). mAb 190 detected only the HIF-2α fusion protein (F) and HIF-2α was absent in the normoxic cell pellet (G), but hypoxically inducible (H). The intensity of nuclear staining observed within the hypoxic cell pellet was heterogeneous for both antigens. Original magnifications, ×100 (A, B, E, and F) and ×400 (C, D, G, and H).
Figure 3.
HIF-1α and HIF-2α expression in many common cancers. Immunostaining demonstrated heterogeneous patterns of HIF-1α and HIF-2α expression within subsets of tumor cells in many common cancers. Panels illustrated are from a breast carcinoma (A and B), a hepatocellular carcinoma (C, positive nuclei marked by arrows, and D), and a hypernephroma (E and F). Original magnifications, ×200 (A−D) and ×400 (E and F).
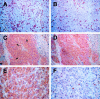
Figure 4.
HIF-1α and HIF-2α expression in breast carcinoma. HIF-1α and HIF-2α protein expression was observed in breast tumor cells adjacent to areas of tumor necrosis. In some cases nuclear expression of HIF-1α was found exclusively at the necrotic/viable tumor margin (A; original magnification, ×100, inset; original magnification, ×200; N, necrosis; S, stroma), but in other cases throughout the tumor (B; original magnification, ×200, inset; original magnification, ×300). Nuclear expression of HIF-2α was observed throughout most tumors (C; original magnification, ×200, D; original magnification, ×400 higher power view of the marked section of the field illustrated in C) though like HIF-1α can also be perinecrotic. There were no fundamental differences between the tissue distributions of HIF-1α and HIF-2α. Serial sections of the case of breast carcinoma illustrated in A, C, and D above were hybridized with 35S-labeled VEGF antisense RNA probe (E, bright field illumination; F, dark field view; original magnifications, ×100) and 35S-labeled sense control probe (G, dark field view; original magnification, ×100). VEGF mRNA is strongly expressed around the large area of intratumoral necrosis (N) where HIF-1α and HIF-2α protein was present.
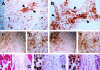
Figure 5.
HIF-2α expression in tumor macrophages. In a variety of tumor types two overlapping distribution patterns were observed with mAb 190b: staining within subsets of the tumor cells and also of stromal cells and macrophages, which was predominantly cytoplasmic. These features are illustrated in sections taken from breast carcinoma. In A, nuclear staining within tumor cells is indicated by arrows and cytoplasmic staining of stromal cells by arrowheads. A further example of stromal staining is shown in B. The immunoreactivity seen in tumor stroma with mAb 190b was investigated further. In serial sections of breast carcinoma colocalization of CD68, using mAb PGM1 (C and D) and mAb 190b (E and F) confirms that the cells staining with 190b in the stroma are macrophages. In serial sections of pancreatic carcinoma, cells shown to be macrophages by their expression of CD68 (detected using mAb PGM1, G) are also labeled by polyclonal antibody PM8 (H) and mAb 190b (I). Because PM8 and 190b recognize different HIF-2α epitopes, this confirms that these macrophages are indeed expressing HIF-2α. Original magnifications, ×200 (A, D, and F−I) and ×100 (B, C, and E)
Figure 6.
HIF-2α mRNA expression by tumor stroma. Serial paraffin sections of a case of breast carcinoma demonstrated to have abundant stromal HIF-2α protein by immunostaining (Figure 5B) ▶ were hybridized with 35S-labeled HIF-2α antisense RNA probe (A, bright field view; B, dark field view), 35S-labeled sense control probe (C, dark field illumination). In contrast, HIF-1α mRNA signal was distributed equally over stromal and tumor areas (D bright field view; E dark field view), 35S-labeled sense control probe (F dark field illumination). Original magnifications, ×100.
Figure 7.
Expression of HIF-2α protein in normal bone marrow macrophages and U937 cell line. Normal bone marrow macrophages show immunoreactivity with mAb 190b. In serial paraffin sections of normal bone marrow trephine staining with CD68 (using mAb PGM1; A), and mAb 190b (B) colocalize in macrophages but not megakaryocytes. U937 cells were cultured with or without PMA (1.6 × 10− M) and incubated in normoxia (N) or 0.1% hypoxia (H) for 4 hours. Whole cell extracts (75 μg) were prepared, separated by SDS-PAGE, transferred onto PVDF membrane, and analyzed with mAb 190b (C). For comparison a hypoxic cell extract of HIF-2α transfected HT1080 cells (50 μg) prepared after culture for 4 hours in 0.1% hypoxia was run in parallel (lane C). The dominant band detected in each case comigrated at the mobility predicted for HIF-2α. In both differentiated and undifferentiated U937 cells the band detected showed hypoxic induction. However, after differentiation the normoxic levels seen were comparable with those seen in hypoxia in undifferentiated cells. U937 cells were cultured with PMA (1.6 × 10− M) to allow differentiation into macrophages, incubated in normoxia or placed in 0.1% hypoxia for 4 hours, and immunostained with mAbs to HIF-2α (190b) and CD11c (KB 90). HIF-2α expression was absent after normoxic culture (D) but detectable after hypoxic culture (E). CD11c expression, confirming macrophage differentiation, was unaffected by normoxic/hypoxic culture (not shown). Original magnifications, ×400 (A and B) and ×300 (D and E)
Similar articles
- The HIF pathway: implications for patterns of gene expression in cancer.
Wykoff CC, Pugh CW, Harris AL, Maxwell PH, Ratcliffe PJ. Wykoff CC, et al. Novartis Found Symp. 2001;240:212-25; discussion 225-31. doi: 10.1002/0470868716.ch15. Novartis Found Symp. 2001. PMID: 11727931 Review. - Biology of hypoxia-inducible factor-2alpha in development and disease.
Patel SA, Simon MC. Patel SA, et al. Cell Death Differ. 2008 Apr;15(4):628-34. doi: 10.1038/cdd.2008.17. Epub 2008 Feb 15. Cell Death Differ. 2008. PMID: 18259197 Free PMC article. Review.
Cited by
- Hypoxia is important in F‑18 FDG accumulation in thecoma‑fibroma tumors on F‑18 FDG PET/CT scans.
Seino H, Ono S, Miura H, Morohashi S, Wu Y, Tsushima F, Takai Y, Kijima H. Seino H, et al. Mol Med Rep. 2016 May;13(5):3821-7. doi: 10.3892/mmr.2016.5016. Epub 2016 Mar 21. Mol Med Rep. 2016. PMID: 27035330 Free PMC article. - Genotyping analysis and ¹⁸FDG uptake in breast cancer patients: a preliminary research.
Bravatà V, Stefano A, Cammarata FP, Minafra L, Russo G, Nicolosi S, Pulizzi S, Gelfi C, Gilardi MC, Messa C. Bravatà V, et al. J Exp Clin Cancer Res. 2013 Apr 30;32(1):23. doi: 10.1186/1756-9966-32-23. J Exp Clin Cancer Res. 2013. PMID: 23631762 Free PMC article. - Hypoxia-dependent regulation of inflammatory pathways in immune cells.
Taylor CT, Doherty G, Fallon PG, Cummins EP. Taylor CT, et al. J Clin Invest. 2016 Oct 3;126(10):3716-3724. doi: 10.1172/JCI84433. Epub 2016 Jul 25. J Clin Invest. 2016. PMID: 27454299 Free PMC article. Review. - Anti-proliferation and anti-invasion effects of diosgenin on gastric cancer BGC-823 cells with HIF-1α shRNAs.
Mao ZJ, Tang QJ, Zhang CA, Qin ZF, Pang B, Wei PK, Liu B, Chou YN. Mao ZJ, et al. Int J Mol Sci. 2012;13(5):6521-6533. doi: 10.3390/ijms13056521. Epub 2012 May 24. Int J Mol Sci. 2012. PMID: 22754381 Free PMC article. - Hypoxia induces IGFBP3 in esophageal squamous cancer cells through HIF-1α-mediated mRNA transcription and continuous protein synthesis.
Natsuizaka M, Naganuma S, Kagawa S, Ohashi S, Ahmadi A, Subramanian H, Chang S, Nakagawa KJ, Ji X, Liebhaber SA, Klein-Szanto AJ, Nakagawa H. Natsuizaka M, et al. FASEB J. 2012 Jun;26(6):2620-30. doi: 10.1096/fj.11-198598. Epub 2012 Mar 13. FASEB J. 2012. PMID: 22415309 Free PMC article.
References
- Bunn HF, Poyton RO: Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev 1996, 76:839-885 - PubMed
- Hockel M, Schlenger K, Hockel S, Aral B, Schaffer U, Vaupel P: Tumor hypoxia in pelvic recurrences of cervical cancer. Int J Cancer 1998, 79:365-369 - PubMed
- Stadler P, Becker A, Feldmann HJ, Hansgen G, Dunst J, Wurschmidt F, Molls M: Influence of the hypoxic subvolume on the survival of patients with head and neck cancer. Int J Radiat Oncol Biol Phys 1999, 44:749-754 - PubMed
- Ratcliffe PJ, Ebert BL, Firth JD, Gleadle JM, Maxwell PH, Nagao M, Orourke JF, Pugh CW, Wood SM: Oxygen regulated gene expression: erythropoietin as a model system. Kidney Int 1997, 51:514-526 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources