The apoptosome: heart and soul of the cell death machine - PubMed (original) (raw)
Review
The apoptosome: heart and soul of the cell death machine
A M Chinnaiyan. Neoplasia. 1999 Apr.
Abstract
Apoptosis is a fundamental biologic process by which metazoan cells orchestrate their own self-demise. Genetic analyses of the nematode C elegans identified three core components of the suicide apparatus which include CED-3, CED-4, and CED-9. An analogous set of core constituents exists in mammalian cells and includes caspase-9, Apaf-1, and bcl-2/xL, respectively. CED-3 and CED-4, along with their mammalian counterparts, function to kill cells, whereas CED-9 and its mammalian equivalents protect cells from death. These central components biochemically intermingle in a ternary complex recently dubbed the "apoptosome." The C elegans protein EGL-1 and its mammalian counterparts, pro-apoptotic members of the bcl-2 family, induce cell death by disrupting apoptosome interactions. Thus, EGL-1 may represent a primordial signal integrator for the apoptosome. Various biochemical processes including oligomerization, adenosine triphosphate ATP/dATP binding, and cytochrome c interaction play a role in regulating the ternary death complex. Recent studies suggest that cell death receptors, such as CD95, may amplify their suicide signal by activating the apoptosome. These mutual associations by core components of the suicide apparatus provide a molecular framework in which diverse death signals likely interface. Understanding the apoptosome and its cellular connections will facilitate the design of novel therapeutic strategies for cancer and other disease states in which apoptosis plays a pivotal role.
Figures
Figure 1
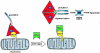
The apoptosome: A molecular framework for cell death C elegans (left) and mammals (right). In C elegans, CED-4 plays a crucial role as a molecular death switch, interacting with and assisting in the activation of the latent death protease CED-3. CED-9 presumably interacts with and negatively regulates CED-4 function. EGL-1 can disrupt the apoptosome by sequestering CED-9. With some added complexity, this primordial paradigm is likely recapitulated in mammals. Unlike C elegans, the mammalian death machinery is complicated by redundancy. Bcl-xl, bcl-2, and other death suppressors of the bcl-2 family inhibit apoptosis by negatively regulating Apaf-1. By contrast, death-promoting members of the bcl-2 family such as bid, bik, and bad antagonize the interactions between bcl-2/xl and Apaf-1. In the absence of negative regulation, Apaf-1 is free to engage and activate the death protease, caspase-9. Cytochrome c and ATP/dATP likely function as cofactors for the apoptosome. Once triggered, apical caspases such as caspase-9 activate downstream small prodomain caspases.
Figure 2
Schematic representation of apoptosome constituents.
Figure 3
The C elegans apoptosome and its mechanism of activation. In the inactive state the ternary complex (CED-3, CED-4, and CED-9) is assembled on mitochondria. An unspecified death signal mobilizes EGL-1. In turn, EGL-1 binds CED-9, disengaging the apoptosome. CED-4 proceeds to oligomerize and induce the proximity and consequent activation of proCED-3 molecules.
Figure 4
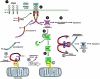
The mammalian apoptosome and its interface with the CD95 pathway. 1: Unactivated CD95(Fas/APO-1) expressed on the cell surface. DD, death domain. DED, death effector domain. 2: Formation of the CD95 death-signaling complex after trimerization by CD95L. Initially, the death adaptor FADD is recruited the cytoplasmic domain of CD95. FADD in turn induces proximity among procaspase-8 molecules, leading to 3: proteolytic activation. 4: Active caspase-8 then cleaves small-prodomain caspases, leading to cell death. 5: Alternatively, caspase-i can cleave the bcl-2 homologue Bid. 6: Once Bid is proteolytically cleaved to tBid (truncated) it can bind bcl-xl. In the inactive state (A), Apaf-1 is in a closed conformation bound to bcl-xl. The asterisk represents the P-loop motif. CARD, caspase recruitment domain. Upon death stimulus, cytochrome c is released from mitochondria (B). Likewise, death signals mobilize proapoptotic members of the bcl-2 family, such as Bid, to disrupt the latent apoptosome (C). Cytochrome c and/or dATP/ATP hydrolysis induces a conformational change in Apaf-1, negating the inhibitory effect of the WD-40 repeats (black boxes in the Apaf-1 molecule). In the open conformation and liberated of bcl-xl inhibition, Apaf-1 is free to self-associate via its CED-4-like domain (D). CARD-CARD associations link Apaf-1 to procaspase-9. Oligomerization of Apaf-1 aggregates procaspase-9, leading to its proteolytic activation (E) and consequent induction of cell death.
Similar articles
- Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death.
Chinnaiyan AM, O'Rourke K, Lane BR, Dixit VM. Chinnaiyan AM, et al. Science. 1997 Feb 21;275(5303):1122-6. doi: 10.1126/science.275.5303.1122. Science. 1997. PMID: 9027312 - Apoptosome assembly.
Shi Y. Shi Y. Methods Enzymol. 2008;442:141-56. doi: 10.1016/S0076-6879(08)01407-9. Methods Enzymol. 2008. PMID: 18662568 - The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9.
Conradt B, Horvitz HR. Conradt B, et al. Cell. 1998 May 15;93(4):519-29. doi: 10.1016/s0092-8674(00)81182-4. Cell. 1998. PMID: 9604928 - Chemical-induced apoptosis: formation of the Apaf-1 apoptosome.
Cain K. Cain K. Drug Metab Rev. 2003 Nov;35(4):337-63. doi: 10.1081/dmr-120026497. Drug Metab Rev. 2003. PMID: 14705865 Review. - [Molecular mechanism of apoptosis].
Shimizu S, Tsujimoto Y. Shimizu S, et al. Seikagaku. 1998 Jan;70(1):14-21. Seikagaku. 1998. PMID: 9503981 Review. Japanese. No abstract available.
Cited by
- Anastatica hierochuntica (L.) methanolic and aqueous extracts exert antiproliferative effects through the induction of apoptosis in MCF-7 breast cancer cells.
Rameshbabu S, Messaoudi SA, Alehaideb ZI, Ali MS, Venktraman A, Alajmi H, Al-Eidi H, Matou-Nasri S. Rameshbabu S, et al. Saudi Pharm J. 2020 Aug;28(8):985-993. doi: 10.1016/j.jsps.2020.06.020. Epub 2020 Jul 9. Saudi Pharm J. 2020. PMID: 32792843 Free PMC article. - The Molecular Links between Cell Death and Inflammasome.
Lee KH, Kang TB. Lee KH, et al. Cells. 2019 Sep 10;8(9):1057. doi: 10.3390/cells8091057. Cells. 2019. PMID: 31509938 Free PMC article. Review. - Increased Oxidative Stress Induced by Rubus Bioactive Compounds Induce Apoptotic Cell Death in Human Breast Cancer Cells.
George BP, Abrahamse H. George BP, et al. Oxid Med Cell Longev. 2019 Jun 3;2019:6797921. doi: 10.1155/2019/6797921. eCollection 2019. Oxid Med Cell Longev. 2019. PMID: 31281587 Free PMC article. - Redox state-dependent aggregation of mitochondria induced by cytochrome c.
Lemeshko VV. Lemeshko VV. Mol Cell Biochem. 2012 Jan;360(1-2):111-9. doi: 10.1007/s11010-011-1049-1. Epub 2011 Sep 9. Mol Cell Biochem. 2012. PMID: 21904946 - ATP and cytochrome c-dependent activation of caspase-9 during hypoxia in the cerebral cortex of newborn piglets.
Delivoria-Papadopoulos M, Gorn M, Ashraf QM, Mishra OP. Delivoria-Papadopoulos M, et al. Neurosci Lett. 2007 Dec 18;429(2-3):115-9. doi: 10.1016/j.neulet.2007.09.072. Epub 2007 Oct 11. Neurosci Lett. 2007. PMID: 17976908 Free PMC article.
References
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1988;396:643–649. - PubMed
- van der Biezen EA, Jones JD. The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol. 1998;8:R226–R227. [letter] - PubMed
- del Pozo O, Lam E. Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr Biol. 1998;8:R896. [In Process Citation] - PubMed
- Chinnaiyan AM, Chaudhary D, O'Rourke K, Koonin EV, Dixit VM. Role of CED-4 in the activation of CED-3. Nature. 1997;388:728–729. [letter] [see comments] - PubMed
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous