Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus - PubMed (original) (raw)
Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus
T Aragón et al. Mol Cell Biol. 2000 Sep.
Abstract
Influenza virus NS1 protein is an RNA-binding protein whose expression alters several posttranscriptional regulatory processes, like polyadenylation, splicing, and nucleocytoplasmic transport of cellular mRNAs. In addition, NS1 protein enhances the translational rate of viral, but not cellular, mRNAs. To characterize this effect, we looked for targets of NS1 influenza virus protein among cellular translation factors. We found that NS1 coimmunoprecipitates with eukaryotic initiation factor 4GI (eIF4GI), the large subunit of the cap-binding complex eIF4F, either in influenza virus-infected cells or in cells transfected with NS1 cDNA. Affinity chromatography studies using a purified His-NS1 protein-containing matrix showed that the fusion protein pulls down endogenous eIF4GI from COS-1 cells and labeled eIF4GI translated in vitro, but not the eIF4E subunit of the eIF4F factor. Similar in vitro binding experiments with eIF4GI deletion mutants indicated that the NS1-binding domain of eIF4GI is located between residues 157 and 550, in a region where no other component of the translational machinery is known to interact. Moreover, using overlay assays and pull-down experiments, we showed that NS1 and eIF4GI proteins interact directly, in an RNA-independent manner. Mapping of the eIF4GI-binding domain in the NS1 protein indicated that the first 113 N-terminal amino acids of the protein, but not the first 81, are sufficient to bind eIF4GI. The first of these mutants has been previously shown to act as a translational enhancer, while the second is defective in this activity. Collectively, these and previously published data suggest a model where NS1 recruits eIF4GI specifically to the 5' untranslated region (5' UTR) of the viral mRNA, allowing for the preferential translation of the influenza virus messengers.
Figures
FIG. 1
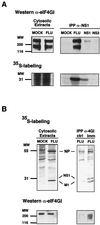
In vivo association of NS1 to eIF4GI. Cultures of COS-1 cells were infected with influenza virus A/Victoria/3/75 (FLU), transfected with plasmid pSVa232NS1 (NS1) or pSVa232NS2 (NS2), or mock transfected (MOCK). The cultures were labeled with [35S]methionine-cysteine (35S-labeling), and cytoplasmic extracts were prepared at 5 h postinfection (A), 8 h postinfection (B), or 60 h posttransfection. (A) Shown are cytosolic extracts immunoprecipitated with anti-NS1 (IPP α-NS1) and the immunoprecipitates and samples of the extracts prior to immunoprecipitation (Cytosolic Extracts), separated in polyacrylamide-SDS gels. The gels were blotted to nitrocellulose and exposed for autoradiography. Finally, the filters were developed in a Western assay using anti-eIF4GI serum. (B) Cytosolic extracts were immunoprecipitated with either preimmune (ctrl) or anti-4GI serum (imm) and separated in polyacrylamide-SDS gels. The gels were dried and exposed for autoradiography (upper panels) or blotted to nitrocellulose and developed for Western assay using anti-eIF4GI serum (lower panels).
FIG. 2
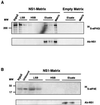
NS1 pulls down initiation factor eIF4GI. Cytoplasmic fractions of COS-1 cells were incubated with a His-NS1-containing matrix (A), a His-PAΔ1-464-containing matrix (B), or an empty matrix (C). After incubation for 1 h, resins were washed with HSB and the retained protein was eluted with 1 M imidazol (Eluate), as described in Materials and Methods. Fractions corresponding to cellular extract (Extract), the last wash (L. wash), and eluate were analyzed by SDS-PAGE, transferred to Immobilon (Millipore), and developed for Western assays with specific anti-eIF4GI, anti-NS1, or anti-HisPx antibodies. Ten percent of the input and 30% of the fractions eluted with 1 M imidazole were analyzed. The left side of panel B shows the purified His-PA deletion-containing protein used in the His-PA-containing matrix.
FIG. 3
NS1 retains in vitro synthesized eIF4GI but not eIF4E. Plasmids peIF-4GIΔ156 and pMV-7 were used in an in vitro transcription-translation reaction in the presence of [35S]methionine-cysteine to generate eIF4GIΔ156 (A) and eIF4E (B) proteins, respectively. Postribosomal supernatants of the translation mixtures were applied to an NS1-containing matrix (NS1-Matrix) or to an empty matrix. After incubation for 1 h, resins were washed with LSB, HSB, and 1 M imidazole (Eluate). Total supernatant (Input), samples of the different fractions, and the remaining resins (Matrix) were analyzed by denaturing gels and exposed for autoradiography. Ten percent of the input and 30% of the other samples are shown in the figure. Fractions used in LSB and HSB represent the first and the last samples of these washing conditions. The same samples were analyzed for the presence of NS1 protein by Western assays with specific antibodies (Ab-NS1).
FIG. 4
Mapping of eIF4GI-interacting domains. (A) Scheme of the eIF4GI regions involved in the interaction with translation-related proteins and of the eIF4GI proteins used in panel B. (B) The N-terminal domain of eIF4GI interacts with NS1. Postribosomal supernatants of in vitro labeled eIF4GI recombinants were applied to an NS1-containing matrix (His-NS1 matrix) or to an empty matrix and incubated for 1 h. After incubation, the resins were extensively washed, and samples of input (Input) or the protein retained by the different matrices were analyzed by SDS-PAGE and exposed for autoradiography.
FIG. 5
NS1 and eIF4GI interact directly. (A) Overlay experiments. The upper left panel shows the purification of His-eIF4GI157-550 and His-NS1 recombinant proteins. The proteins were purified by Ni2+-containing resins as described in Materials and Methods, analyzed by SDS-polyacrylamide gels, and stained with Coomassie blue. The upper right panel indicates the specificity of the antibodies. A total of 500 ng of protein A, BSA, purified His-eIF4GI157-550 (4GI157-550), or His-NS1 (NS1) protein was applied to nitrocellulose filters and developed with antibodies specific for NS1 or eIF4GI or with an anti-rabbit peroxidase for protein A detection. The lower panel represents an overlay assay. Different amounts (from 50 to 500 ng) of BSA or purified recombinant His-eIF4GI157-550 (4GI157-550) or His-NS1 (NS1) protein were applied to nitrocellulose filters (Protein bound). The filters were incubated with solutions containing either protein A, His-eIF4GI157-550, or His-NS1 at 5 ng/ml, were fixed with formaldehyde, and were extensively washed (Probed with). Finally, the binding of the probe protein was detected by Western blotting. Where indicated, both proteins used for binding were treated with RNase A. (B) Pull-down experiments. The left panel shows GST or GST-NS1 expressed and purified from bacteria as described in Materials and Methods. The right panel indicates His-eIF4GI157-550 protein incubated with GST- or GST-NS1-containing resin. After incubation and extensive washing, the retained protein was eluted with 10 mM glutathione. Samples of purified His-eIF4Gt157-550 (Input), unbound protein (Unb), eluate, and the corresponding matrix were analyzed by SDS-PAGE followed by Western assays with antibodies against His (Ab-HisPx) or GST (Ab-GST).
FIG. 6
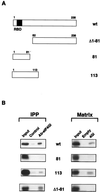
Mapping of eIF4GI-binding domain in the NS1 protein. (A) Scheme of the NS1 deletion mutants. (B) Mapping of interacting domains. Total extracts of in vitro-labeled full length NS1 (wt) and deletion mutant proteins were coimmunoprecipitated (IPP) with a control serum (Control) or with a specific anti-eIF4GI serum (Ab-eIF4GI). Alternatively, in vitro translated NS1 proteins were used for pull-down experiments (Matrix) with an empty matrix or with a matrix containing His-4GI157-550 (4GI). Samples of input (Input), of the coimmunoprecipitates, or of the protein retained by the different matrices were analyzed by SDS-PAGE and exposed for autoradiography.
FIG. 7
Composition of eIF4F complex in influenza virus-infected cells. Cultures of COS-1 cells were mock infected (MOCK) or infected with influenza virus A/Victoria/3/75 (FLU). The cultures were labeled with [35S]Met-Cys, and cytoplasmic extracts were prepared at 6 h postinfection. (A) Cytosolic extracts were separated in SDS-polyacrylamide gels and exposed for autoradiography. (B) Cytosolic extracts were immunoprecipitated with preimmune (IPP α-4GI, ctrl) or immune (IPP α-4GI, imm) anti-4GI sera. Samples of the immunoprecipitates or the extracts prior to immunoprecipitation (Cytosolic Extracts) were separated in SDS-polyacrylamide gels. The gels were blotted to nitrocellulose and developed in a Western assay using anti-eIF4GI (Ab-eIF4GI), anti-eIF4E (Ab-eIF4E), or anti-PABP (Ab-PABP) serum.
Similar articles
- PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes.
Burgui I, Aragón T, Ortín J, Nieto A. Burgui I, et al. J Gen Virol. 2003 Dec;84(Pt 12):3263-3274. doi: 10.1099/vir.0.19487-0. J Gen Virol. 2003. PMID: 14645908 - Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons.
Byrd MP, Zamora M, Lloyd RE. Byrd MP, et al. Mol Cell Biol. 2002 Jul;22(13):4499-511. doi: 10.1128/MCB.22.13.4499-4511.2002. Mol Cell Biol. 2002. PMID: 12052860 Free PMC article. - Strategies of Influenza A Virus to Ensure the Translation of Viral mRNAs.
Li HC, Yang CH, Lo SY. Li HC, et al. Pathogens. 2022 Dec 12;11(12):1521. doi: 10.3390/pathogens11121521. Pathogens. 2022. PMID: 36558855 Free PMC article. Review. - Translational control of viral gene expression in eukaryotes.
Gale M Jr, Tan SL, Katze MG. Gale M Jr, et al. Microbiol Mol Biol Rev. 2000 Jun;64(2):239-80. doi: 10.1128/MMBR.64.2.239-280.2000. Microbiol Mol Biol Rev. 2000. PMID: 10839817 Free PMC article. Review.
Cited by
- The NS1 protein of influenza A virus interacts with cellular processing bodies and stress granules through RNA-associated protein 55 (RAP55) during virus infection.
Mok BW, Song W, Wang P, Tai H, Chen Y, Zheng M, Wen X, Lau SY, Wu WL, Matsumoto K, Yuen KY, Chen H. Mok BW, et al. J Virol. 2012 Dec;86(23):12695-707. doi: 10.1128/JVI.00647-12. Epub 2012 Sep 12. J Virol. 2012. PMID: 22973032 Free PMC article. - Surveillance in Eastern India (2007-2009) revealed reassortment event involving NS and PB1-F2 gene segments among co-circulating influenza A subtypes.
Sarkar M, Chanda S, Chakrabarti S, Mazumdar J, Ganguly A, Chadha MS, Mishra AC, Chawla-Sarkar M. Sarkar M, et al. Virol J. 2012 Jan 5;9:3. doi: 10.1186/1743-422X-9-3. Virol J. 2012. PMID: 22217077 Free PMC article. - Structural and functional analysis of NS1 and NS2 proteins of H1N1 subtype.
Salahuddin P, Khan AU. Salahuddin P, et al. Genomics Proteomics Bioinformatics. 2010 Sep;8(3):190-9. doi: 10.1016/S1672-0229(10)60021-6. Genomics Proteomics Bioinformatics. 2010. PMID: 20970747 Free PMC article. - The NS1 protein of a human influenza virus inhibits type I interferon production and the induction of antiviral responses in primary human dendritic and respiratory epithelial cells.
Haye K, Burmakina S, Moran T, García-Sastre A, Fernandez-Sesma A. Haye K, et al. J Virol. 2009 Jul;83(13):6849-62. doi: 10.1128/JVI.02323-08. Epub 2009 Apr 29. J Virol. 2009. PMID: 19403682 Free PMC article. - Evolution of Therapeutic Antibodies, Influenza Virus Biology, Influenza, and Influenza Immunotherapy.
Chaisri U, Chaicumpa W. Chaisri U, et al. Biomed Res Int. 2018 May 28;2018:9747549. doi: 10.1155/2018/9747549. eCollection 2018. Biomed Res Int. 2018. PMID: 29998138 Free PMC article. Review.
References
- Beloso A, Martínez C, Valcárcel J, Fernández-Santarén J, Ortín J. Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff. J Gen Virol. 1992;73:575–581. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous