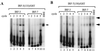Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7 - PubMed (original) (raw)
Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7
R Lin et al. Mol Cell Biol. 2000 Sep.
Abstract
Recent studies implicate the interferon (IFN) regulatory factors (IRF) IRF-3 and IRF-7 as key activators of the alpha/beta IFN (IFN-alpha/beta) genes as well as the RANTES chemokine gene. Using coexpression analysis, the human IFNB, IFNA1, and RANTES promoters were stimulated by IRF-3 coexpression, whereas the IFNA4, IFNA7, and IFNA14 promoters were preferentially induced by IRF-7 only. Chimeric proteins containing combinations of different IRF-7 and IRF-3 domains were also tested, and the results provided evidence of distinct DNA binding properties of IRF-3 and IRF-7, as well as a preferential association of IRF-3 with the CREB binding protein (CBP) coactivator. Interestingly, some of these fusion proteins led to supraphysiological levels of IFN promoter activation. DNA binding site selection studies demonstrated that IRF-3 and IRF-7 bound to the 5'-GAAANNGAAANN-3' consensus motif found in many virus-inducible genes; however, a single nucleotide substitution in either of the GAAA half-site motifs eliminated IRF-3 binding and transactivation activity but did not affect IRF-7 interaction or transactivation activity. These studies demonstrate that IRF-3 possesses a restricted DNA binding site specificity and interacts with CBP, whereas IRF-7 has a broader DNA binding specificity that contributes to its capacity to stimulate delayed-type IFN gene expression. These results provide an explanation for the differential regulation of IFN-alpha/beta gene expression by IRF-3 and IRF-7 and suggest that these factors have complementary rather than redundant roles in the activation of the IFN-alpha/beta genes.
Figures
FIG. 1
Activities of IRF-7 and IRF-3 fusion proteins. The structures of the fusion proteins are illustrated schematically (Pro, proline-rich domain; TAO, transactivation domain; SRD, signal response domain; CAD, constitutive activation domain; ID, inhibitory domain). IRF-7/3A contains 541 amino acids, 246 from IRF-7 (1 to 246) and 295 from IRF-3(5D) (133 to 427). IRF-7/3B consists of 495 amino acids, 200 from IRF-7 (1 to 200) and 295 from IRF-3(5D) (133 to 427). IRF-7/3C consists of 445 amino acids, 150 from IRF-7 (1 to 150) and 295 from IRF-3(5D) (133 to 427). IRF-3/7 contains 265 amino acids, 132 from IRF-3 and 133 from IRF-7(Δ247–467). For transactivation assays, 293 cells were transfected with the pRLTK control plasmid, the RANTES-pGL3, IFNB-pGL3, IFNA1-pGL3, or IFNA4-pGL3 reporter plasmid, and the expression plasmids encoding IRF-3, IRF-3(5D), IRF-7, IRF-7(D477/479), IRF-7(Δ247–467), IRF-3/7, or IRF-7/3, as indicated. Luciferase activity was analyzed at 24 h posttransfection by the dual-luciferase reporter assay as described by the manufacturer (Promega). Relative luciferase activity was measured as fold activation (relative to the basal level for the reporter gene in the presence of the pFlag-CMV-2 vector after normalization to cotransfected relative light unit activity); the values represent the average of three experiments performed in duplicate, with variability of 10 to 25%.
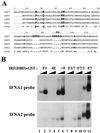
FIG. 2
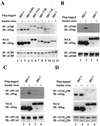
DNA binding activity of IRF-7/3 chimeric proteins. (A) An EMSA was performed with whole-cell extracts (20 μg) derived from 293 cells transfected with various Flag-tagged IRF-7 or IRF-7/3 expression plasmids. At 24 h posttransfection, cells were infected with Sendai virus for 6 h (+) or left uninfected (−), as indicated. The 32P-labeled probe corresponded to the PRDI-PRDIII (5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′) motif of the IFNB promoter. (B) Twenty micrograms of whole-cell extracts from panel A was analyzed by immunoblotting (IB) with anti-Flag antibody. F7(2D), F7 D477/479.
FIG. 3
Activated forms of IRF-3 and IRF-7/3 chimeric proteins but not IRF-7 are associated with histone acetyltransferases. 293 cells were transfected with Flag-tagged IRF-3, IRF-7, or IRF-7/3 expression plasmids, as indicated above the lanes. At 24 h posttransfection, cells were infected with Sendai virus for 12 h (+) or left uninfected (−), as indicated. Whole-cell extracts (WCE) (200 μg for CBP and 500 μg for p300, PCAF, and TAFIII250) were immunoprecipitated with anti-CBP antibody A-22 (A), anti-p300 antibody N-15 (B), anti-PCAF antibody (C), or anti-TAFIIp250 antibody 6B3 (D). Immunoprecipitated complexes (upper panel) or 20 μg of whole-cell extracts (middle panel) was analyzed by SDS–8% PAGE and subsequently probed with anti-Flag antibody M2. The membranes shown in the upper panel were reprobed with anti-CBP antibody A-22 (A), anti-p300 antibody N-15 (B), anti-PCAF antibody (C), or anti-TAFIIp250 antibody 6B3 (D) (lower panel). IP, immunoprecipitation; IB, immunoblotting.
FIG. 4
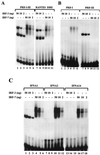
Binding of IRF-3 and IRF-7 to IRF binding sites of the IFN and RANTES promoters. (A) Recombinant N-terminal IRF-3 and IRF-7 bind to PRDI-PRDIII and RANTES ISRE probes. An EMSA was performed with the indicated amounts of recombinant protein; the 32P-labeled probe corresponded to the PRDI-PRDIII region (5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′) or the ISRE of the RANTES gene (5′-CTATTTCAGTTTTCTTTTCCGTTTTGTG-3′). (B) Recombinant N-terminal IRF-3 and IRF-7 bind to PRDI and PRDIII probes. An EMSA was performed with the indicated amounts of recombinant protein; the 32P-labeled probe corresponded to PRDI (5′-GAGAAGTGAAAGTG-3′) or PRDIII (5′-GAAAACTGAAAGGG-3′). (C) Recombinant N-terminal IRF-3 and IRF-7 bind to the PRDI-like and TG sites from the IFNA1, IFNA2, and IFNA14 promoters. An EMSA was performed with the indicated amounts of IRF-3 or IRF-7 and 32P-labeled probes corresponding to the following PRDI-like sites: IFNA1, 5′-GGAAAGCAAAAACAGAAATGGAAAGTGG-3′; IFNA2, 5′-GAAAGCAAAAAGAGAAGTAGAAAGTAA-3′; and IFNA14, 5′-GGAAAGCCAAAAGAGAAGTAGAAAAAAA-3′.
FIG. 5
Gel shift analysis of selected IRF-3 and IRF-7 binding sites. Oligonucleotides selected with recombinant IRF-3–GST (lanes 1 to 5) or IRF-7–GST (lanes 6 to 10) at each round were amplified by PCR using 32P-labeled primers and subsequently used as probes in a gel shift analysis. Fifty nanograms of IRF-3–GST (A) or IRF-7–GST (B) was used in each binding reaction. The number of selection cycles is shown above each lane. Arrows indicate protein-DNA complexes.
FIG. 6
Consensus sequences binding to IRF-3. Sequences of 16 cloned IRF-3 binding sites derived by five rounds of binding site selection are shown. Each sequence was aligned with respect to its homologous sequence (in boldface type). The frequency of each nucleotide at each position of the homologous sequence and the consensus sequence are shown at the bottom.
FIG. 7
Consensus sequences binding to IRF-7. Sequences of 28 cloned IRF-7 binding sites derived by five rounds of binding site selection are shown. Each sequence was aligned with respect to its homologous sequence (in boldface type). The frequency of each nucleotide at each position of the homologous sequence and the consensus sequence are shown at the bottom.
FIG. 8
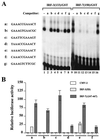
Characterization of selected binding sites. (A) An EMSA was performed with 20 ng of recombinant IRF-3–GST (lanes 1 to 8) or IRF-7–GST (lanes 9 to 16), 32P-labeled oligonucleotide a (5′-GAAACCGAAACTGAAACCGAAACT-3′), and a 1,000-fold molar excess of competitor DNA. Selected binding sites (two copies, indicated beside the gel as a to g) were used as competitors. (B) Activation of selected promoters by IRF-3 and IRF-7. 293 cells were transfected with the pRLTK control plasmid, reporter constructs containing the minimum TK-luciferase promoter and two copies of selected binding sites (designated a to f), and the active forms of IRF-3(5D) and IRF-7(Δ247–467) expression plasmids, and luciferase activity was analyzed at 24 h posttransfection. Relative luciferase activity was measured as fold activation (relative to the basal level for the reporter gene in the presence of the pFlag-CMV-2 vector after normalization to cotransfected relative light unit activity); the values represent the average of three experiments performed in duplicate, with variability of 10 to 25%.
FIG. 9
Amino acids of IRF-7 involved in different DNA binding specificities. (A) Sequence alignment of the DBDs of IRF-7, IRF-3, IRF-2, and IRF-1. The sequential numbering of IRF-7 is shown at the top. Identical residues in IRF-1, IRF-2, and IRF-3 but not IRF-7 are shown in boldface type. (B) Binding of mutated forms of IRF-3 or IRF3 and IRF7 chimeric recombinant proteins to the PRDI-like- and TG sites from the IFNA1 and IFNA2 promoters. An EMSA was performed with the indicated amounts of recombinant GST fusion proteins and 32P-labeled probes corresponding to the following PRDI-like sites: IFNA1, 5′-GGAAAGCAAAAACAGAAATGGAAAGTGG-3′; and IFNA2, 5′-GAAAGCAAAAAGAGAAGTAGAAAGTAA-3′.
Similar articles
- Regulation of type I interferon gene expression by interferon regulatory factor-3.
Schafer SL, Lin R, Moore PA, Hiscott J, Pitha PM. Schafer SL, et al. J Biol Chem. 1998 Jan 30;273(5):2714-20. doi: 10.1074/jbc.273.5.2714. J Biol Chem. 1998. PMID: 9446577 - Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA.
Servant MJ, Grandvaux N, tenOever BR, Duguay D, Lin R, Hiscott J. Servant MJ, et al. J Biol Chem. 2003 Mar 14;278(11):9441-7. doi: 10.1074/jbc.M209851200. Epub 2003 Jan 10. J Biol Chem. 2003. PMID: 12524442 - The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness.
Masumi A, Wang IM, Lefebvre B, Yang XJ, Nakatani Y, Ozato K. Masumi A, et al. Mol Cell Biol. 1999 Mar;19(3):1810-20. doi: 10.1128/MCB.19.3.1810. Mol Cell Biol. 1999. PMID: 10022868 Free PMC article. - On the role of IRF in host defense.
Barnes B, Lubyova B, Pitha PM. Barnes B, et al. J Interferon Cytokine Res. 2002 Jan;22(1):59-71. doi: 10.1089/107999002753452665. J Interferon Cytokine Res. 2002. PMID: 11846976 Review. - Triggering the interferon response: the role of IRF-3 transcription factor.
Hiscott J, Pitha P, Genin P, Nguyen H, Heylbroeck C, Mamane Y, Algarte M, Lin R. Hiscott J, et al. J Interferon Cytokine Res. 1999 Jan;19(1):1-13. doi: 10.1089/107999099314360. J Interferon Cytokine Res. 1999. PMID: 10048763 Review.
Cited by
- Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage.
Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, Hiscott J. Lin R, et al. J Virol. 2006 Jun;80(12):6072-83. doi: 10.1128/JVI.02495-05. J Virol. 2006. PMID: 16731946 Free PMC article. - Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer.
Panne D, Maniatis T, Harrison SC. Panne D, et al. EMBO J. 2004 Nov 10;23(22):4384-93. doi: 10.1038/sj.emboj.7600453. Epub 2004 Oct 28. EMBO J. 2004. PMID: 15510218 Free PMC article. - Transcription factor redundancy ensures induction of the antiviral state.
Schmid S, Mordstein M, Kochs G, García-Sastre A, Tenoever BR. Schmid S, et al. J Biol Chem. 2010 Dec 31;285(53):42013-22. doi: 10.1074/jbc.M110.165936. Epub 2010 Oct 13. J Biol Chem. 2010. PMID: 20943654 Free PMC article. - P200 family protein IFI204 negatively regulates type I interferon responses by targeting IRF7 in nucleus.
Cao L, Ji Y, Zeng L, Liu Q, Zhang Z, Guo S, Guo X, Tong Y, Zhao X, Li CM, Chen Y, Guo D. Cao L, et al. PLoS Pathog. 2019 Oct 11;15(10):e1008079. doi: 10.1371/journal.ppat.1008079. eCollection 2019 Oct. PLoS Pathog. 2019. PMID: 31603949 Free PMC article. - RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation.
Widau RC, Parekh AD, Ranck MC, Golden DW, Kumar KA, Sood RF, Pitroda SP, Liao Z, Huang X, Darga TE, Xu D, Huang L, Andrade J, Roizman B, Weichselbaum RR, Khodarev NN. Widau RC, et al. Proc Natl Acad Sci U S A. 2014 Jan 28;111(4):E484-91. doi: 10.1073/pnas.1323253111. Epub 2014 Jan 13. Proc Natl Acad Sci U S A. 2014. PMID: 24434553 Free PMC article.
References
- Au W C, Moore P A, LaFleur D W, Tombal B, Pitha P M. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J Biol Chem. 1998;273:29210–29217. - PubMed
- Braganca J, Civas A. Type I interferon gene expression: differential expression of IFN-A genes induced by viruses and double-stranded RNA. Biochimie. 1998;80:673–687. - PubMed
- Burysek L, Yeow W S, Pitha P M. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2:19–32. - PubMed
- Chen J, Attardi L, Verrijzer P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources