Multiple splicing defects in an intronic false exon - PubMed (original) (raw)
Multiple splicing defects in an intronic false exon
H Sun et al. Mol Cell Biol. 2000 Sep.
Abstract
Splice site consensus sequences alone are insufficient to dictate the recognition of real constitutive splice sites within the typically large transcripts of higher eukaryotes, and large numbers of pseudoexons flanked by pseudosplice sites with good matches to the consensus sequences can be easily designated. In an attempt to identify elements that prevent pseudoexon splicing, we have systematically altered known splicing signals, as well as immediately adjacent flanking sequences, of an arbitrarily chosen pseudoexon from intron 1 of the human hprt gene. The substitution of a 5' splice site that perfectly matches the 5' consensus combined with mutation to match the CAG/G sequence of the 3' consensus failed to get this model pseudoexon included as the central exon in a dhfr minigene context. Provision of a real 3' splice site and a consensus 5' splice site and removal of an upstream inhibitory sequence were necessary and sufficient to confer splicing on the pseudoexon. This activated context also supported the splicing of a second pseudoexon sequence containing no apparent enhancer. Thus, both the 5' splice site sequence and the polypyrimidine tract of the pseudoexon are defective despite their good agreement with the consensus. On the other hand, the pseudoexon body did not exert a negative influence on splicing. The introduction into the pseudoexon of a sequence selected for binding to ASF/SF2 or its replacement with beta-globin exon 2 only partially reversed the effect of the upstream negative element and the defective polypyrimidine tract. These results support the idea that exon-bridging enhancers are not a prerequisite for constitutive exon definition and suggest that intrinsically defective splice sites and negative elements play important roles in distinguishing the real splicing signal from the vast number of false splicing signals.
Figures
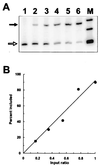
FIG. 1
Quantitation of the RT-PCR assay for exon skipping. An RNA preparation from a cell population exhibiting complete pseudoexon skipping (DH2) was mixed in various ratios with an RNA preparation from a cell population exhibiting mostly (90%) pseudoexon inclusion (DH3t3). The sample amounts were chosen so that the unmixed samples (lanes 1 and 6) produced signals similar in intensity. After RT-PCR, the radioactive PCR products were separated by polyacrylamide gel electrophoresis. (A) Phosphorimage of the PCR products. The closed arrow indicates exon inclusion, and the open arrow indicates exon skipping. Lanes 1 to 6 represent mixtures containing the RNA exhibiting 90% exon inclusion in the following proportions of the total: 0, 0.17, 0.35, 0.55, 0.76, and 1, respectively. Lane M, φX174 _Hin_fI fragments. (B) Graphical representation of the data in panel A.
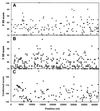
FIG. 2
Pseudosplice sites and pseudoexons in the human hprt gene. (A) Locations and consensus scores of 9-nt sequences resembling 5′ splice sites in the human hprt gene. The open symbols indicate the eight real 5′ splice sites (SS). Only those sites having scores equal to or higher than the lowest real 5′ splice site scores (75 for intron 3) are shown. (B) Locations and consensus scores of 15-nt sequences resembling 3′ splice sites. The open symbols indicate the eight real 5′ splice sites. Only those 285 sites having scores equal to or higher than 75 are shown. The lowest scores of real 3′ splice site are 71 and 69 for introns 6 and 7, respectively; these two points are plotted as 75 and indicated by downward-pointing open triangles. There are 675 sequences that would meet the lower cutoff of 69. (C) Locations and combined 3′ and 5′ scores of 103 pseudoexons in the hprt gene. The criteria for a pseudoexon were a 3′ pseudosplice site scoring at least 69, followed within at least 50 nt but no more than 200 nt by a 5′ pseudosplice site scoring at least 75, plus the presence of a sequence resembling a branch point within 60 nt upstream of the 3′ pseudosplice site (see Materials and Methods). If more than one pseudoexon shared a 3′ or 5′ pseudosplice site, only the highest-scoring candidate was chosen. The arrow points to the score of pseudoexon 1, which was selected for further study.
FIG. 3
Sequence changes at the 3′ pseudosplice site of hprt pseudoexon 1. The slash indicates the predicted potential 3′ splice site. Upstream dhfr host minigene sequences are in italics. Point mutations are in lowercase. The intronic splicing silencer 34-mer and its truncated and mutated versions are underlined. An inverted 34-mer sequence is overlined. A 10-nt PPT taken from the authentic 3′ splice site of hprt intron 1 is shaded. The original 5′ splice site sequence downstream of the pseudoexon was CAG/GUGUGA, which is present here only in pDH2. All of the other constructs in this list contained the 5′ splice site consensus sequence CAG/GUGaGu.
FIG. 4
Splicing characteristics of constructs carrying various configurations of pseudoexon 1. (A) Schematic representations of pseudoexon 1 constructs. Individual constructs are described in the text. Ψ, hprt pseudoexon 1; S, mutation of a single nonsense codon to sense in the body of the pseudoexon (all constructs other than pDH1 contain this change); ∗, mutation of the original CAG/C sequence at the 3′ pseudosplice site to the consensus CAG/G; ∗∗, mutation of the original CAGG/GUGUGA sequence at the 5′ pseudosplice site to the consensus CAG/GUGAGU; X and A, _Xho_I and _Apa_I restriction sites, respectively. (B and C) Phosphorimages of RT-PCR products from total RNA extracted from permanently transfected cells. The closed arrow indicates exon inclusion (I) and the open arrow indicates exon skipping (S) in dhfr minigene transcripts; closed arrow 1, 2, or 3 indicates inclusion of the inserted exon in the construct pD2P2, pDH2, or pD2B, respectively. The closed arrowhead indicates inclusion and the open arrowhead indicates skipping of pseudoexon 1 in aprt transcripts. The markers (lanes M) were φX174 _Hin_fI fragments. U, unspliced transcripts.
FIG. 5
Effect of altering the upstream flank and 3′ and 5′ splice sites on the splicing of pseudoexon 1. (A) Schematic representations of pseudoexon constructs. Individual constructs are described in the text. The asterisks represent consensus CAG/G and CAG/GUGAGU sequences as described in the legend to Fig. 4. An open bar indicates a sequence derived from the 3′ end of hprt intron 1 (an authentic 3′ splice site), except for pAD, where it was derived from dhfr intron 1. The 34r in construct pDH4dt denotes the reverse sequence of the 34-mer intronic splicing silencer. The tc in the diagram of pDH3pM denotes a GU-to-TC mutation that removes a potential competing GU dinucleotide at position +7. In the column labeled Inclusion, +means >90% inclusion of the central exon, − means skipping of the central exon, and a number indicates percent inclusion of the central exon (percent included/included + skipped). (B, C, D, and E) Phosphorimages of RT-PCR products from total RNA extracted from permanently transfected cells (B, C, and D) or transiently transfected cells (E). The closed arrows indicate exon inclusion and the open arrows indicate exon skipping in the dhfr minigene transcripts. The closed arrowhead indicates inclusion of aprt exon 4 in the pAPRTSS2 context. Lanes M, φX174 _Hin_fI fragments.
FIG. 6
Effect of exon body sequence alteration on splicing. (A) Schematic representations of pseudoexon constructs and derivatives. Individual constructs are described in the text. A single asterisk indicates mutation to the CAG/G of a consensus 3′ splice site, and a double asterisk indicates mutation to a perfect CAG/GUGAGU 5′ consensus splice site. A3 denotes a 72-nt insert containing three tandem repeats of an ASF/SF2-binding SELEX winning sequence, and S3 denotes an analogous insert containing three tandem repeats of an SC35-binding SELEX winning sequence (84). Gex2 indicates human β-globin exon 2, and Ψ-2 indicates hprt pseudoexon 2. (B and C) Phosphorimages of RT-PCR products of RNA extracted from permanently transfected cells. The closed arrows indicate exon inclusion and the open arrows indicate exon skipping in the dhfr minigene context. Lanes M, φX174 _Hin_fI fragments. Closed arrows in panel C indicate the following: 1, size of unspliced RNA (or DNA) in constructs containing the globin exon; 2, size of RNA that has retained intron 1 in pDG12D (confirmed by sequencing); 3, size of RNA produced by splicing of the central exon at an upstream cryptic 3′ splice site in pDG12 and pDG12D (confirmed by sequencing); 4, size corresponding to inclusion of β-globin exon 2 in pDG11 (confirmed by sequencing); 5, size corresponding to inclusion of pseudoexon 2 in pDHP2; 6, size corresponding to skipping of the central exon in all constructs of the dhfr minigene. Lanes M, φX174 _Hin_fI fragments.
Similar articles
- Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells.
Carothers AM, Urlaub G, Grunberger D, Chasin LA. Carothers AM, et al. Mol Cell Biol. 1993 Aug;13(8):5085-98. doi: 10.1128/mcb.13.8.5085-5098.1993. Mol Cell Biol. 1993. PMID: 8336736 Free PMC article. - An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing.
Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F. Zuccato E, et al. J Biol Chem. 2004 Apr 23;279(17):16980-8. doi: 10.1074/jbc.M313439200. Epub 2004 Feb 13. J Biol Chem. 2004. PMID: 14966131 - Pseudoexon activation in disease by non-splice site deep intronic sequence variation - wild type pseudoexons constitute high-risk sites in the human genome.
Petersen USS, Doktor TK, Andresen BS. Petersen USS, et al. Hum Mutat. 2022 Feb;43(2):103-127. doi: 10.1002/humu.24306. Epub 2021 Dec 5. Hum Mutat. 2022. PMID: 34837434 Review. - Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum.
O'Neill JP, Rogan PK, Cariello N, Nicklas JA. O'Neill JP, et al. Mutat Res. 1998 Nov;411(3):179-214. doi: 10.1016/s1383-5742(98)00013-1. Mutat Res. 1998. PMID: 9804951 Review.
Cited by
- hnRNPM protects against the dsRNA-mediated interferon response by repressing LINE-associated cryptic splicing.
Zheng R, Dunlap M, Bobkov GOM, Gonzalez-Figueroa C, Patel KJ, Lyu J, Harvey SE, Chan TW, Quinones-Valdez G, Choudhury M, Le Roux CA, Bartels MD, Vuong A, Flynn RA, Chang HY, Van Nostrand EL, Xiao X, Cheng C. Zheng R, et al. Mol Cell. 2024 Jun 6;84(11):2087-2103.e8. doi: 10.1016/j.molcel.2024.05.004. Epub 2024 May 29. Mol Cell. 2024. PMID: 38815579 - Long non-coding RNAs: roles in cellular stress responses and epigenetic mechanisms regulating chromatin.
Nickerson JA, Momen-Heravi F. Nickerson JA, et al. Nucleus. 2024 Dec;15(1):2350180. doi: 10.1080/19491034.2024.2350180. Epub 2024 May 22. Nucleus. 2024. PMID: 38773934 Free PMC article. Review. - Successful skipping of abnormal pseudoexon by antisense oligonucleotides in vitro for a patient with beta-propeller protein-associated neurodegeneration.
Yamada M, Maeta K, Suzuki H, Kurosawa R, Takenouchi T, Awaya T, Ajiro M, Takeuchi A, Nishio H, Hagiwara M, Miya F, Matsuo M, Kosaki K. Yamada M, et al. Sci Rep. 2024 Mar 18;14(1):6506. doi: 10.1038/s41598-024-56704-z. Sci Rep. 2024. PMID: 38499569 Free PMC article. - Introns and Their Therapeutic Applications in Biomedical Researches.
Haddad-Mashadrizeh A, Mirahmadi M, Taghavizadeh Yazdi ME, Gholampour-Faroji N, Bahrami A, Zomorodipour A, Moghadam Matin M, Qayoomian M, Saebnia N. Haddad-Mashadrizeh A, et al. Iran J Biotechnol. 2023 Oct 1;21(4):e3316. doi: 10.30498/ijb.2023.334488.3316. eCollection 2023 Oct. Iran J Biotechnol. 2023. PMID: 38269198 Free PMC article. Review. - Introns: the "dark matter" of the eukaryotic genome.
Girardini KN, Olthof AM, Kanadia RN. Girardini KN, et al. Front Genet. 2023 May 16;14:1150212. doi: 10.3389/fgene.2023.1150212. eCollection 2023. Front Genet. 2023. PMID: 37260773 Free PMC article. Review.
References
- Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques. 1995;18:746–750. - PubMed
- Balvay L, Libri D, Fiszman M Y. Pre-mRNA secondary structure and the regulation of splicing. Bioessays. 1993;15:165–169. - PubMed
- Batzer M A, Deininger P L, Hellmann-Blumberg U, Jurka J, Labuda D, Rubin C M, Schmid C W, Zietkiewicz E, Zuckerkandl E. Standardized nomenclature for Alu repeats. J Mol Evol. 1996;42:3–6. - PubMed
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous