Structure-function analysis of a lupus anti-DNA autoantibody: central role of the heavy chain complementarity-determining region 3 Arg in binding of double- and single-stranded DNA - PubMed (original) (raw)
Structure-function analysis of a lupus anti-DNA autoantibody: central role of the heavy chain complementarity-determining region 3 Arg in binding of double- and single-stranded DNA
Z Li et al. Eur J Immunol. 2000 Jul.
Abstract
To determine the contribution of the somatic point mutations and that of the complementarity-determining region (CDR)3 Arg to DNA binding, we engineered the germline V(H) and V(kappa) gene revertant and site-mutagenized the CDR3 Arg residues of the mutated and "antigen-selected" mAb 412.67. This anti-DNA autoantibody was derived from B-1 cells of a lupus patient and bore two H-CDR3 Arg, Arg105 and Arg107, encoded by N segment additions, and one kappa-CDR3 Arg, Arg97, resulting from a point mutation (Kasaian et al. 1994. J. Immunol. 152: 3137-3151; Kasaian et al. 1995. Ann. N.Y Acad. Sci. 764: 410-423). The germ-line revertant bound double-stranded (ds) DNA and single-stranded (ss) DNA as effectively as its wild-type counterpart (relative avidity: 6.4x10(-7) and 9.9x10(-9) vs. 6.7x10(-7) and 9.1 x10(-9) g/microl), raising the possibility that an antigen other than DNA was responsible for the selection of the mAb 412.67 V(H) and V(kappa) point mutations. H-CDR3 Arg105 and Arg107 were both required for dsDNA binding, but either Arg105 or Arg107 was sufficient for ssDNA binding. The central role of Arg105 and Arg107 in DNA binding reflected their solvent-exposed orientation at the apex of the H-CDR3 main loop. Consistent with its inward orientation afar from the antigen-binding surface, the kappa-CDR3 Arg97 played no role in either dsDNA or ssDNA binding.
Figures
Fig. 1
Nucleotide sequences of the wild-type, variously substituted, and germ-line (top row) mAb 412.67 VHDJH and VκJκ gene segments. 412.67.gl.5a is the autologous germ-line gene sequence. Dashes indicate identities. Solid lines on top of each cluster depict CDR. Lowercase letters denote vector sequences. The sequences or reverse complementary sequences encompassed by the H-Ov1, H-Ov2, H-FR3, 67cFR3, H-FR4, 67-CDR3 A, and 67-CDR3 B or κ-Ov1, κ-Ov2, κIII-FR3 A, κIII-FR3 B, and κ-FR4 mutagenized primers are underlined.
Fig. 2
Deduced amino acid sequences of the wild-type, variously substituted, and germ-line (top row) mAb 412.67 VHDJH and VκJκ gene segments. 412.67.gl.5a is the autologous germ-line sequence. Dashes indicate identities. Solid lines on top of each cluster depict CDR.
Fig. 3
Generation of the germ-line revertant and mutagenized mAb 412.67 VHDJH and VκJκ DNA, their insertion into the pcDNAIG and the pSXRDIG vectors and expression in F3B6 cells.
Fig. 4
Binding of the original IgA1κ mAb 412.67 (A), recombinant “wild-type” VH67-D(R105, R107)JH67/Vκ67-(R97) Jκ67 (B), “germ-line” VH67gl.5a-D(R105, R107) JH67/VκKv325(R97)Jκ67 (C), “wild-type CDR3 Gly-substituted” VH67-D(G105, G107)JH67/Vκ67-(W97)Jκ67 (D), “Trp-CDR3” revertant VH67-D (R105, R107) JH67/Vκ67(W97)Jκ67 (E), “wild-type CDR3 Gly-substituted” VH67-D(G105, G107) JH67/Vκ67(R97)Jκ67 (F), “single CDR3 Gly-substituted” VH67gl.5a-D(G105, R107)JH67/Vκ67(R97)Jκ67 (G), “single CDR3 Gly-substituted” VH67gl.5a-D(R105, G107)JH67/Vκ67(R97)Jκ67 (H) IgG1κ to ssDNA (■) and dsDNA (□). Bars schematize the composition of original wild-type IgA1κ, the wild-type IgG1κ, and the various recombinant and mutagenized molecules (black rectangles depict the wild-type sequences derived from the original mAb 412.67; white rectangles depict the gene sequence derived from autologous germ-line gene segments or the H-CDR3 Gly-substituted sequences). Antigen-binding activity is expressed as absorbance at 492 nm.
Fig. 5
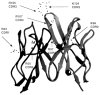
Ribbon drawing of mAb 412.67 Fv showing the κ chain on the left and the H chain on the right. All Arg and Lys in the H-CDR and κ-CDR are indicated.
Fig. 6
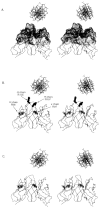
Stereo images of α-carbon trace drawings of the modeled mAb 412.67 Fv with B form DNA positioned above the binding site, drawn to scale. The molecular surface covering the CDR residues is depicted as dots (A). The H-CDR3 Arg 105 (R105) and 107 (R107) are depicted as full circles; the κ-CDR3 Arg97 (R97) and the H-FR3 Arg98 (R98) are depicted as open circles (B). The H-CDR3 Arg105 and Arg107 are substituted with Gly residues (C).
Similar articles
- Chromatin specificity of anti-double-stranded DNA antibodies and a role for Arg residues in the third complementarity-determining region of the heavy chain.
Guth AM, Zhang X, Smith D, Detanico T, Wysocki LJ. Guth AM, et al. J Immunol. 2003 Dec 1;171(11):6260-6. doi: 10.4049/jimmunol.171.11.6260. J Immunol. 2003. PMID: 14634143 - Genetic, structural and functional properties of an IgG DNA-binding monoclonal antibody from a lupus patient with nephritis.
Ravirajan CT, Rahman MA, Papadaki L, Griffiths MH, Kalsi J, Martin AC, Ehrenstein MR, Latchman DS, Isenberg DA. Ravirajan CT, et al. Eur J Immunol. 1998 Jan;28(1):339-50. doi: 10.1002/(SICI)1521-4141(199801)28:01<339::AID-IMMU339>3.0.CO;2-C. Eur J Immunol. 1998. PMID: 9485213 - Genetic and structural evidence for antigen selection of anti-DNA antibodies.
Radic MZ, Weigert M. Radic MZ, et al. Annu Rev Immunol. 1994;12:487-520. doi: 10.1146/annurev.iy.12.040194.002415. Annu Rev Immunol. 1994. PMID: 8011289 Review. - Origins of anti-DNA antibodies and their implications for B-cell tolerance.
Radic MZ, Weigert M. Radic MZ, et al. Ann N Y Acad Sci. 1995 Sep 29;764:384-96. doi: 10.1111/j.1749-6632.1995.tb55853.x. Ann N Y Acad Sci. 1995. PMID: 7486552 Review. No abstract available.
Cited by
- Molecular and structural basis of anti-DNA antibody specificity for pyrrolated proteins.
Anan Y, Itakura M, Shimoda T, Yamaguchi K, Lu P, Nagata K, Dong J, Ueda H, Uchida K. Anan Y, et al. Commun Biol. 2024 Feb 3;7(1):149. doi: 10.1038/s42003-024-05851-0. Commun Biol. 2024. PMID: 38310133 Free PMC article. - A Human MSH6 Germline Variant Associated With Systemic Lupus Erythematosus Induces Lupus-like Disease in Mice.
Meas R, Nititham J, Taylor KE, Maher S, Clairmont K, Carufe KEW, Kashgarian M, Nottoli T, Cheong A, Nagel ZD, Gaffney PM, Criswell LA, Sweasy JB. Meas R, et al. ACR Open Rheumatol. 2022 Sep;4(9):760-770. doi: 10.1002/acr2.11471. Epub 2022 Jun 16. ACR Open Rheumatol. 2022. PMID: 35708944 Free PMC article. - Heavy chain-only antibody genes in fish evolved to generate unique CDR3 repertoire.
Anumukonda K, Francis M, Currie P, Tulenko F, Hsu E. Anumukonda K, et al. Eur J Immunol. 2022 Feb;52(2):247-260. doi: 10.1002/eji.202149588. Epub 2021 Nov 19. Eur J Immunol. 2022. PMID: 34708869 Free PMC article. - Autoantibody-dependent amplification of inflammation in SLE.
Lou H, Wojciak-Stothard B, Ruseva MM, Cook HT, Kelleher P, Pickering MC, Mongkolsapaya J, Screaton GR, Xu XN. Lou H, et al. Cell Death Dis. 2020 Sep 9;11(9):729. doi: 10.1038/s41419-020-02928-6. Cell Death Dis. 2020. PMID: 32908129 Free PMC article. - Antibody Epitope Specificity for dsDNA Phosphate Backbone Is an Intrinsic Property of the Heavy Chain Variable Germline Gene Segment Used.
Srdic-Rajic T, Kohler H, Jurisic V, Metlas R. Srdic-Rajic T, et al. Front Immunol. 2018 Oct 18;9:2378. doi: 10.3389/fimmu.2018.02378. eCollection 2018. Front Immunol. 2018. PMID: 30405605 Free PMC article.
References
- Zan H, Li Z-D, Dramitinos P, Schaffer A, Cerutti A, Casali P. BCR engagement and T cell contact induce bcl-6 somatic hypermutation in human B cells: association with initiation of transcription and identity with Ig hypermutation. J Immunol. 2000 in press. - PubMed
- Diamond B, Katz JB, Paul E, Aranow C, Lustgarten D, Scharff MD. The role of somatic mutation in the pathogenic anti-DNA response. Annu Rev Immunol. 1992;10:731–757. - PubMed
- Radic MZ, Weigert M. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu Rev Immunol. 1994;12:487–520. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous