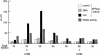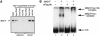dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties - PubMed (original) (raw)
dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties
A Brehm et al. EMBO J. 2000.
Abstract
Mi-2 and ISWI, two members of the Snf2 superfamily of ATPases, reside in separate ATP-dependent chromatin remodelling complexes. These complexes differ in their biochemical properties and are believed to perform distinct functions in the cell. We have compared the remodelling activity of recombinant Drosophila Mi-2 (dMi-2) with that of recombinant ISWI. Both proteins are nucleosome-stimulated ATPases and promote nucleosome mobilization. However, dMi-2 and ISWI differ in their interaction with nucleosome core particles, in their substrate requirements and in the direction of nucleosome mobilization. We have used antibodies to immobilize a complex containing dMi-2 and the dRPD3 histone deacetylase from Drosophila embryo extracts. This complex shares the nucleosome-stimulated ATPase and nucleosome mobilization properties of recombinant dMi-2, demonstrating that these activities are maintained in a physiological context. Its functional properties distinguish dMi-2 from both SWI2/SNF2 and ISWI, defining a new family of ATP-dependent remodelling machines.
Figures
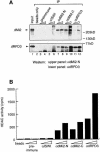
Fig. 1. (A) dMi-2 and dRPD3 coimmunoprecipitate. Drosophila embryo nuclear extract was immunoprecipitated with 1 and 5 µl of various antisera as indicated. dMi-2 and dRPD3 were detected by western analysis using the αdMi-2-N (upper panel) or the αdRPD3 antiserum (lower panel). Lane 1: 20% input. Positions of dMi-2 and dRPD3 bands are indicated on the left, molecular weights are indicated on the right. (B) αdMi-2 antibodies coprecipitate histone deacetylase activity. Immunoprecipitations were performed as in (A) using 1 and 5 µl of antisera. HDAC activity was measured using a radioactive histone H4 peptide as a substrate. (C) αdMi-2 and αdRPD3 antibodies coprecipitate nucleosome-stimulated ATPase activity. Immunoprecipitations were performed as in (A). Antisera used are indicated at the top. Each immunoprecipitate was split into three and used for ATPase assays in the absence or presence of 50 ng of free or nucleosomal DNA. ATPase activity values are expressed as percentage conversion of ATP.
Fig. 1. (A) dMi-2 and dRPD3 coimmunoprecipitate. Drosophila embryo nuclear extract was immunoprecipitated with 1 and 5 µl of various antisera as indicated. dMi-2 and dRPD3 were detected by western analysis using the αdMi-2-N (upper panel) or the αdRPD3 antiserum (lower panel). Lane 1: 20% input. Positions of dMi-2 and dRPD3 bands are indicated on the left, molecular weights are indicated on the right. (B) αdMi-2 antibodies coprecipitate histone deacetylase activity. Immunoprecipitations were performed as in (A) using 1 and 5 µl of antisera. HDAC activity was measured using a radioactive histone H4 peptide as a substrate. (C) αdMi-2 and αdRPD3 antibodies coprecipitate nucleosome-stimulated ATPase activity. Immunoprecipitations were performed as in (A). Antisera used are indicated at the top. Each immunoprecipitate was split into three and used for ATPase assays in the absence or presence of 50 ng of free or nucleosomal DNA. ATPase activity values are expressed as percentage conversion of ATP.
Fig. 2. dMi-2 resides in a large complex. Partially purified dMi-2-containing fractions (see Materials and methods) were fractionated over a Superose 6 sizing column. Aliquots of fractions were immunoprecipitated with 1 µl of αdMi-2-C and subjected to ATPase assays, or immunoprecipitated with 3 µl of αdRPD3 antiserum and subjected to the HDAC assay as indicated. Molecular weights of fractions eluting from the Superose 6 column were determined with molecular weight standards (Pharmacia) and are indicated below fraction numbers.
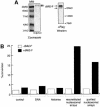
Fig. 3. Recombinant dMi-2 is a nucleosome-stimulated ATPase. (A) Recombinant dMi-2-F (0.3 pmol) was applied to SDS–PAGE and either stained with Coomassie Blue (left panel) or analysed by western blotting using α-Flag antibody (right panel). The position of dMi-2-F-derived bands is indicated by arrows. MW: molecular weight marker. (B) Recombinant dMi-2-F (200 fmol) was used for ATPase assays in the absence (control) or presence of 50 ng of naked DNA (DNA), 50 ng of core histones purified from Drosophila embryos (histones), 50 ng of nucleosomal arrays reconstituted by salt gradient dialysis (reconstituted nucleosomal arrays) and 50 ng of nucleosomal arrays purified from Drosophila embryos as indicated.
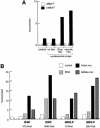
Fig. 4. Activation of dMi-2 by recombinant nucleosomes. (A) Recombinant dMi-2-F (200 fmol) was used for ATPase assays in the absence (control) or presence of 50 ng of nucleosomal arrays assembled with yNAP1 as a histone deposition vehicle. To the yNAP1 assembly reactions either no histones (no hist.), histones purified from Drosophila embryos (Dros. hist.) or recombinant histones expressed in E.coli (recomb. hist.) were added. (B and C) Nucleosomal arrays were assembled with yNAP1 using either no histones (DNA), recombinant intact histones (intact nuc) or recombinant histones lacking all four tails (tailless nuc) and used to stimulate ATPase activities of recombinant ISWI and dMi-2-F as indicated. The amounts of ISWI and dMi-2-F in (B) were chosen to give similar activation by intact nucleosomes. ATPase assay and quantification were done as described above.
Fig. 4. Activation of dMi-2 by recombinant nucleosomes. (A) Recombinant dMi-2-F (200 fmol) was used for ATPase assays in the absence (control) or presence of 50 ng of nucleosomal arrays assembled with yNAP1 as a histone deposition vehicle. To the yNAP1 assembly reactions either no histones (no hist.), histones purified from Drosophila embryos (Dros. hist.) or recombinant histones expressed in E.coli (recomb. hist.) were added. (B and C) Nucleosomal arrays were assembled with yNAP1 using either no histones (DNA), recombinant intact histones (intact nuc) or recombinant histones lacking all four tails (tailless nuc) and used to stimulate ATPase activities of recombinant ISWI and dMi-2-F as indicated. The amounts of ISWI and dMi-2-F in (B) were chosen to give similar activation by intact nucleosomes. ATPase assay and quantification were done as described above.
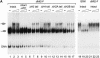
Fig. 5. Recombinant dMi-2 binds to nucleosomes. (A) Biotinylated linearized DNA was bound to streptavidin-coated paramagnetic beads and assembled into nucleosomal arrays using yNAP1 as a histone deposition vehicle. Assembly reactions contained either no histones (free DNA), Drosophila histones (Dros.hist.) or recombinant histones (rec.hist.) as indicated. Bound and unbound materials were subjected to SDS–PAGE and western blot analysis using the M2 antibody (anti-Flag; Sigma). Thirty percent input was loaded as a control (input). (B) Recombinant dMi-2-F (240 fmol, lanes 2 and 4) was incubated with mononucleosomes containing a 248 bp radioactively labelled DNA fragment in the absence or presence of α-Flag antibody as shown. Resulting complexes were separated by native gel electrophoresis and visualized following autoradiography. Free mononucleosomes (nuc.) and dMi-2-F′–nucleosome complexes are indicated by arrows. (C) Recombinant dMi-2-F (lanes 2 and 8: 40 fmol; lanes 3 and 9: 80 fmol; lanes 4 and 10: 120 fmol; lanes 5 and 11: 240 fmol; lane 6: 360 fmol) and ISWI (lanes 13 and 20: 5 fmol; lanes 14 and 21: 10 fmol; lanes 15 and 22: 15 fmol; lanes 16 and 23: 25 fmol; lanes 17 and 24: 50 fmol; lanes 18 and 25: 75 fmol) were incubated with mononucleosomes containing 146 or 248 bp of radioactively labelled DNA as indicated. The positions of the 146 bp (grey ellipse) and 248 bp nucleosome (grey ellipse with protruding DNA ends) are shown on the left. The positions of nucleosome complexes are indicated by arrows.
Fig. 5. Recombinant dMi-2 binds to nucleosomes. (A) Biotinylated linearized DNA was bound to streptavidin-coated paramagnetic beads and assembled into nucleosomal arrays using yNAP1 as a histone deposition vehicle. Assembly reactions contained either no histones (free DNA), Drosophila histones (Dros.hist.) or recombinant histones (rec.hist.) as indicated. Bound and unbound materials were subjected to SDS–PAGE and western blot analysis using the M2 antibody (anti-Flag; Sigma). Thirty percent input was loaded as a control (input). (B) Recombinant dMi-2-F (240 fmol, lanes 2 and 4) was incubated with mononucleosomes containing a 248 bp radioactively labelled DNA fragment in the absence or presence of α-Flag antibody as shown. Resulting complexes were separated by native gel electrophoresis and visualized following autoradiography. Free mononucleosomes (nuc.) and dMi-2-F′–nucleosome complexes are indicated by arrows. (C) Recombinant dMi-2-F (lanes 2 and 8: 40 fmol; lanes 3 and 9: 80 fmol; lanes 4 and 10: 120 fmol; lanes 5 and 11: 240 fmol; lane 6: 360 fmol) and ISWI (lanes 13 and 20: 5 fmol; lanes 14 and 21: 10 fmol; lanes 15 and 22: 15 fmol; lanes 16 and 23: 25 fmol; lanes 17 and 24: 50 fmol; lanes 18 and 25: 75 fmol) were incubated with mononucleosomes containing 146 or 248 bp of radioactively labelled DNA as indicated. The positions of the 146 bp (grey ellipse) and 248 bp nucleosome (grey ellipse with protruding DNA ends) are shown on the left. The positions of nucleosome complexes are indicated by arrows.
Fig. 6. dMi-2 mobilizes mononucleosomes. (A) Recombinant dMi-2 and ISWI were incubated with positioned mononucleosomes reconstituted with either four wild-type recombinant histones (intact) or three wild-type histones and one lacking the N-terminal tail (Δ) as indicated. Nucleosomes were then separated by native gel electrophoresis. The positions of free DNA, end-positioned mononucleosomes and centre-positioned mononucleosomes are shown on the left. (B) Recombinant dMi-2 and immunoprecipitates (see Materials and methods) were incubated with positioned nucleosomes as described above. The antisera used and presence of ATP in the reactions are indicated on the top (beads: no antibody control). rec.dMi2-F: recombinant dMi-2-F. The positions of free DNA and the two nucleosome species are shown on the left.
Fig. 6. dMi-2 mobilizes mononucleosomes. (A) Recombinant dMi-2 and ISWI were incubated with positioned mononucleosomes reconstituted with either four wild-type recombinant histones (intact) or three wild-type histones and one lacking the N-terminal tail (Δ) as indicated. Nucleosomes were then separated by native gel electrophoresis. The positions of free DNA, end-positioned mononucleosomes and centre-positioned mononucleosomes are shown on the left. (B) Recombinant dMi-2 and immunoprecipitates (see Materials and methods) were incubated with positioned nucleosomes as described above. The antisera used and presence of ATP in the reactions are indicated on the top (beads: no antibody control). rec.dMi2-F: recombinant dMi-2-F. The positions of free DNA and the two nucleosome species are shown on the left.
Similar articles
- The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization.
Bouazoune K, Mitterweger A, Längst G, Imhof A, Akhtar A, Becker PB, Brehm A. Bouazoune K, et al. EMBO J. 2002 May 15;21(10):2430-40. doi: 10.1093/emboj/21.10.2430. EMBO J. 2002. PMID: 12006495 Free PMC article. - Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI.
Clapier CR, Längst G, Corona DF, Becker PB, Nightingale KP. Clapier CR, et al. Mol Cell Biol. 2001 Feb;21(3):875-83. doi: 10.1128/MCB.21.3.875-883.2001. Mol Cell Biol. 2001. PMID: 11154274 Free PMC article. - Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC).
Corona DF, Eberharter A, Budde A, Deuring R, Ferrari S, Varga-Weisz P, Wilm M, Tamkun J, Becker PB. Corona DF, et al. EMBO J. 2000 Jun 15;19(12):3049-59. doi: 10.1093/emboj/19.12.3049. EMBO J. 2000. PMID: 10856248 Free PMC article. - ATP-dependent nucleosome remodeling.
Becker PB, Hörz W. Becker PB, et al. Annu Rev Biochem. 2002;71:247-73. doi: 10.1146/annurev.biochem.71.110601.135400. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045097 Review. - Chromatin remodelling in mammalian cells by ISWI-type complexes--where, when and why?
Erdel F, Rippe K. Erdel F, et al. FEBS J. 2011 Oct;278(19):3608-18. doi: 10.1111/j.1742-4658.2011.08282.x. Epub 2011 Sep 2. FEBS J. 2011. PMID: 21810179 Review.
Cited by
- Energy-driven genome regulation by ATP-dependent chromatin remodellers.
Eustermann S, Patel AB, Hopfner KP, He Y, Korber P. Eustermann S, et al. Nat Rev Mol Cell Biol. 2024 Apr;25(4):309-332. doi: 10.1038/s41580-023-00683-y. Epub 2023 Dec 11. Nat Rev Mol Cell Biol. 2024. PMID: 38081975 Review. - DNA sequence encoded repression of rRNA gene transcription in chromatin.
Felle M, Exler JH, Merkl R, Dachauer K, Brehm A, Grummt I, Längst G. Felle M, et al. Nucleic Acids Res. 2010 Sep;38(16):5304-14. doi: 10.1093/nar/gkq263. Epub 2010 Apr 25. Nucleic Acids Res. 2010. PMID: 20421213 Free PMC article. - NuRD-independent Mi-2 activity represses ectopic gene expression during neuronal maturation.
Aughey GN, Forsberg E, Grimes K, Zhang S, Southall TD. Aughey GN, et al. EMBO Rep. 2023 Apr 5;24(4):e55362. doi: 10.15252/embr.202255362. Epub 2023 Feb 1. EMBO Rep. 2023. PMID: 36722816 Free PMC article. - Developmental roles of the Mi-2/NURD-associated protein p66 in Drosophila.
Kon C, Cadigan KM, da Silva SL, Nusse R. Kon C, et al. Genetics. 2005 Apr;169(4):2087-100. doi: 10.1534/genetics.104.034595. Epub 2005 Feb 3. Genetics. 2005. PMID: 15695365 Free PMC article. - The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization.
Bouazoune K, Mitterweger A, Längst G, Imhof A, Akhtar A, Becker PB, Brehm A. Bouazoune K, et al. EMBO J. 2002 May 15;21(10):2430-40. doi: 10.1093/emboj/21.10.2430. EMBO J. 2002. PMID: 12006495 Free PMC article.
References
- Akthar A. and Becker,P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell, 5, 1–9. - PubMed
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. - PubMed
- Cairns B.R. (1998) Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci., 23, 20–25. - PubMed
- Corona D.F., Langst,G., Clapier,C.R., Bonte,E.J., Ferrari,S., Tamkun,J.W. and Becker,P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases