Detection and characterization of genetic recombination in cytopathic type 2 bovine viral diarrhea viruses - PubMed (original) (raw)
Detection and characterization of genetic recombination in cytopathic type 2 bovine viral diarrhea viruses
J F Ridpath et al. J Virol. 2000 Sep.
Abstract
In cytopathic bovine viral diarrhea virus genotype 1 (BVDV1) isolates, insertions are reported at position A (amino acid [aa] 1535) and position B (aa 1589). Insertions at position B predominate. In this survey it was found that in BVDV2, insertions at position A predominate. Possible reasons for this difference in relative frequency are discussed.
Figures
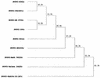
FIG. 1
Phylogenetic analysis of deduced amino acid sequences flanking the NS2/NS3 junction. Consensus amino acid sequences of the NS2/NS3 junction region minus insertion sequences were deduced from sequences generated by using primers shown in Table 1. Sequences compared corresponded to amino acid residues 1290 to 1610 in BVDV1-SD-1. The dendrogram was produced by applying the unpaired geometric mean analysis method using the Higgins-Sharp algorithm (CLUSTAL4) supplied in the MacDNASIS software package (Hitachi Software). Calculated matching percentages are indicated at each branch point of the dendrogram.
FIG. 2
Insertion sites in the NS2-3 coding region. (A) Arrows show the sites of reported insertions into cytopathic BVDV1. Position A corresponds to amino acid residue 1535 and position B corresponds to amino acid residue 1589 of BVDV-1 strain SD-1 (GenBank accession number M96751). (B) The NS2-3 coding regions derived from nine cytopathic BVDV2 minus inserted sequences are shown. One noncytopathic virus coisolated with a cytopathic virus (BVDV2-296nc) and one noncytopathic BVDV2 sequence from GenBank (BVDV2-890, GenBank accession number BVU18059) were included for comparison purposes. Arrows indicate sites of insertions. No arrow indicates that there was no insertion. Unlike their noncytopathic counterparts, eight of these viruses had genomic insertions. No insertion was detected in BVDV2-96B3491c. One virus, BVDV2-MsSt T4529c, had an insertion at position B. Four viruses, BVDV2-ND 8799c, BVDV2-Galena 16425c, BVDV2-Ok St 94-050-297c, and BVDV2-SD1630c, had insertions at position A. Three viruses, BVDV2-296c, BVDV2-5912c, and BVDV2-6082c, had insertions within 10 amino acid residues of position A.
FIG. 3
Inserted sequences detected in cytopathic BVDV2. (A) The insertion in BVDV2-MsSt T4529c at position B consisted of two full-length ubiquitin genes and a partial ubiquitin gene (34 amino acid residues from the amino acid terminus are missing). (B) Seven viruses had insertions at or within 10 amino acid residues of position A. Each of these insertions contained a portion of a cellular gene that codes for a novel DnaJ-like protein (underlined sequences). For three of these viruses, BVDV2-Galena 16452c, BVDV2-OkSt 94-050-297c, and BVDV2-6082c, the DnaJ-like sequence was proceeded by 5, 22, and 28 amino acid residues, respectively. For virus BVDV2-ND 8799c, the DnaJ-like sequence was followed by a string of 10 amino acid residues.
Similar articles
- Delayed onset postvaccinal mucosal disease as a result of genetic recombination between genotype 1 and genotype 2 BVDV.
Ridpath JF, Bolin SR. Ridpath JF, et al. Virology. 1995 Sep 10;212(1):259-62. doi: 10.1006/viro.1995.1480. Virology. 1995. PMID: 7676642 - Genomic analyses of bovine viral diarrhea viruses isolated from cattle imported into Japan between 1991 and 2005.
Yamamoto T, Kozasa T, Aoki H, Sekiguchi H, Morino S, Nakamura S. Yamamoto T, et al. Vet Microbiol. 2008 Mar 18;127(3-4):386-91. doi: 10.1016/j.vetmic.2007.08.020. Epub 2007 Aug 19. Vet Microbiol. 2008. PMID: 17881158 - Natural recombination in bovine viral diarrhea viruses.
Ridpath JF, Qi F, Bolin SR, Berry ES. Ridpath JF, et al. Arch Virol Suppl. 1994;9:239-44. doi: 10.1007/978-3-7091-9326-6_24. Arch Virol Suppl. 1994. PMID: 8032255 - Differentiation of types 1a, 1b and 2 bovine viral diarrhoea virus (BVDV) by PCR.
Ridpath JF, Bolin SR. Ridpath JF, et al. Mol Cell Probes. 1998 Apr;12(2):101-6. doi: 10.1006/mcpr.1998.0158. Mol Cell Probes. 1998. PMID: 9633045
Cited by
- Compartmentalized evolution of Bovine Viral Diarrhoea Virus type 2 in an immunotolerant persistently infected cow.
Colitti B, Nogarol C, Giacobini M, Capucchio MT, Biasato I, Rosati S, Bertolotti L. Colitti B, et al. Sci Rep. 2019 Oct 29;9(1):15460. doi: 10.1038/s41598-019-52023-w. Sci Rep. 2019. PMID: 31664116 Free PMC article. - Molecular chaperone Jiv promotes the RNA replication of classical swine fever virus.
Guo K, Li H, Tan X, Wu M, Lv Q, Liu W, Zhang Y. Guo K, et al. Virus Genes. 2017 Jun;53(3):426-433. doi: 10.1007/s11262-017-1448-9. Epub 2017 Mar 24. Virus Genes. 2017. PMID: 28341934 - Complete Genome Sequence of Bovine Viral Diarrhea Virus 2 Japanese Reference and Vaccine Strain KZ-91CP.
Sato A, Kameyama K, Nagai M, Tateishi K, Ohmori K, Todaka R, Katayama K, Mizutani T, Yamakawa M, Shirai J. Sato A, et al. Genome Announc. 2015 Feb 12;3(1):e01573-14. doi: 10.1128/genomeA.01573-14. Genome Announc. 2015. PMID: 25676770 Free PMC article. - Molecular characterization of Bovine virus diarrhea viruses species 2 (BVDV-2) from cattle in Turkey.
Oguzoglu TC, Muz D, Yilmaz V, Alkan F, Akça Y, Burgu I. Oguzoglu TC, et al. Trop Anim Health Prod. 2010 Aug;42(6):1175-80. doi: 10.1007/s11250-010-9544-z. Epub 2010 Mar 13. Trop Anim Health Prod. 2010. PMID: 20225008
References
- Collett M S, Larson R, Belzer S K, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genomic organization of a pestivirus. Virology. 1988;165:200–208. - PubMed
- Donis R O, Dubovi E J. Differences in virus-induced polypeptides in cells infected by cytopathic and noncytopathic biotypes of bovine virus diarrhea-mucosal disease virus. Virology. 1987;158:168–173. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources