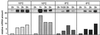Changes in cspL, cspP, and cspC mRNA abundance as a function of cold shock and growth phase in Lactobacillus plantarum - PubMed (original) (raw)
Comparative Study
Changes in cspL, cspP, and cspC mRNA abundance as a function of cold shock and growth phase in Lactobacillus plantarum
S Derzelle et al. J Bacteriol. 2000 Sep.
Abstract
An inverse PCR strategy based on degenerate primers has been used to identify new genes of the cold shock protein family in Lactobacillus plantarum. In addition to the two previously reported cspL and cspP genes, a third gene, cspC, has been cloned and characterized. All three genes encode small 66-amino-acid proteins with between 73 and 88% identity. Comparative Northern blot analyses showed that the level of cspL mRNA increases up to 17-fold after a temperature downshift, whereas the mRNA levels of cspC and cspP remain unchanged or increase only slightly (about two- to threefold). Cold induction of cspL mRNA is transient and delayed in time as a function of the severity of the temperature downshift. The cold shock behavior of the three csp mRNAs contrasts with that observed for four unrelated non-csp genes, which all showed a sharp decrease in mRNA level, followed in one case (bglH) by a progressive recovery of the transcript during prolonged cold exposure. Abundance of the three csp mRNAs was also found to vary during growth at optimal temperature (28 degrees C). cspC and cspP mRNA levels are maximal during the lag period, whereas the abundance of the cspL transcript is highest during late-exponential-phase growth. The differential expression of the three L. plantarum csp genes can be related to sequence and structural differences in their untranslated regions. It also supports the view that the gene products fulfill separate and specific functions, under both cold shock and non-cold shock conditions.
Figures
FIG. 1
Comparison of L. plantarum CspC, CspL, and CspP sequences. Amino acids residues that are identical in all three or just two Csp proteins are shaded in dark and light grey, respectively. The RNA-binding motifs RNP1 and RNP2 are boxed. Conserved residues that are critical for forming the hydrophobic core of the protein (asterisks) or for nucleic acid binding (dots) are indicated.
FIG. 2
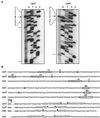
(A) Primer extension analysis of the cspC (left) and cspP (right) transcripts. RNA samples were extracted from cold-shocked cultures (8°C). Bold and thin arrows indicate the positions of major and minor extension products, respectively, obtained with independent primers. Additional signals are nonspecific products that are detected only with individual primers. (B) Nucleotide sequences of the 3′ and 5′ regions of L. plantarum NC8 cspC, cspL, and cspP genes. Only the first and last codons of the Csp-coding sequence are shown. Transcription starts (+1) are circled, as are the 5′ ends of shorter and less abundant primer extension products detected for cspC and cspP. The promoter −35 and −10 boxes and the Shine-Dalgarno sequences (SD) are boxed and circled, respectively. Horizontal arrows indicate the positions of putative transcription terminators.
FIG. 3
Comparative Northern blot analysis of csp and non-csp genes upon cold shock. Exponentially growing cultures of L. plantarum NC8 (OD600 = 0.8) were transferred from 28°C to 8°C, and RNA was prepared at the indicated times before (0 h) and after the temperature downshift. Equal amounts of each RNA sample were run on 1.2% (wt/vol) agarose–0.6% (wt/vol) formaldehyde gels and hybridized with specific radioactive probes for cspC (A), cspP (B), cspL (C), and the four non-csp genes alr, bglH, cbh, and lldh (D). The same membranes were rehybridized with the different probes. One representative result is shown for each gene. Relative mRNA amounts were calculated from the radioactivity measured in the transcript bands at each time point with respect to that found at 0 h. cspL quantification was performed by summing the radioactivities of the short and long transcripts. Data presented for cspC, cspL, and cspP are from three independent experiments.
FIG. 4
Delayed cold induction of cspL mRNA level with decreasing temperatures. RNA was extracted from an exponentially growing NC8 culture (OD600 = 0.8) before and after different times following a temperature downshift from 28°C to 15, 10, 8, or 6°C. The resulting RNA samples were analyzed by hybridization with a cspL probe as described for Fig. 3. As the smaller transcript was barely detectable, quantification was performed only on the large transcript.
FIG. 5
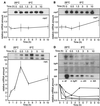
(A) Growth phase-dependent expression of cspC, cspL, and cspP mRNAs. Stationary-phase NC8 cells were diluted with fresh MRS medium and allowed to grow for 25 h at 28°C. RNA samples were taken at different times of culture and analyzed by Northern blot hybridization with cspC, cspP, and cspL probes, as indicated. Results obtained after hybridization of the same membrane with the three different probes are shown. Relative mRNA amount is expressed with respect to the highest level of each transcript during the culture. Radioactivities in the short and large cspL transcripts were summed. (B) Increase of cspC, cspP, and cspL mRNA levels in cold-shocked stationary-phase cells. Relative amounts of cspC, cspP, and cspL transcripts were examined 2 h after transferring stationary-phase NC8 cultures (OD600 = 7) from 28°C to 10°C.
Similar articles
- Cloning and characterization of cspL and cspP, two cold-inducible genes from Lactobacillus plantarum.
Mayo B, Derzelle S, Fernández M, Léonard C, Ferain T, Hols P, Suárez JE, Delcour J. Mayo B, et al. J Bacteriol. 1997 May;179(9):3039-42. doi: 10.1128/jb.179.9.3039-3042.1997. J Bacteriol. 1997. PMID: 9139925 Free PMC article. - Cold stress improves the ability of Lactobacillus plantarum L67 to survive freezing.
Song S, Bae DW, Lim K, Griffiths MW, Oh S. Song S, et al. Int J Food Microbiol. 2014 Nov 17;191:135-43. doi: 10.1016/j.ijfoodmicro.2014.09.017. Epub 2014 Sep 18. Int J Food Microbiol. 2014. PMID: 25261832 - csp-like genes of Lactobacillus delbrueckii ssp. bulgaricus and their response to cold shock.
Serror P, Dervyn R, Ehrlich SD, Maguin E. Serror P, et al. FEMS Microbiol Lett. 2003 Sep 26;226(2):323-30. doi: 10.1016/S0378-1097(03)00594-9. FEMS Microbiol Lett. 2003. PMID: 14553929 - The cold-shock response--a hot topic.
Jones PG, Inouye M. Jones PG, et al. Mol Microbiol. 1994 Mar;11(5):811-8. doi: 10.1111/j.1365-2958.1994.tb00359.x. Mol Microbiol. 1994. PMID: 8022259 Review. - Involvement of CspC in response to diverse environmental stressors in Escherichia coli.
Cardoza E, Singh H. Cardoza E, et al. J Appl Microbiol. 2022 Feb;132(2):785-801. doi: 10.1111/jam.15219. Epub 2021 Aug 21. J Appl Microbiol. 2022. PMID: 34260797 Review.
Cited by
- Development of a Quinoa-Based Fermentation Medium for Propagation of Lactobacillus Plantarum and Weissella Confusa in Opaque Beer Production.
Manhokwe S, Musarurwa T, Jombo TZ, Mugadza DT, Mugari A, Bare J, Mguni S, Chigondo F, Muchekeza JT. Manhokwe S, et al. Int J Microbiol. 2025 Jan 21;2025:5745539. doi: 10.1155/ijm/5745539. eCollection 2025. Int J Microbiol. 2025. PMID: 39963294 Free PMC article. - D-alanyl ester depletion of teichoic acids in Lactobacillus plantarum results in a major modification of lipoteichoic acid composition and cell wall perforations at the septum mediated by the Acm2 autolysin.
Palumbo E, Deghorain M, Cocconcelli PS, Kleerebezem M, Geyer A, Hartung T, Morath S, Hols P. Palumbo E, et al. J Bacteriol. 2006 May;188(10):3709-15. doi: 10.1128/JB.188.10.3709-3715.2006. J Bacteriol. 2006. PMID: 16672624 Free PMC article. - Whole-genome sequencing of Pseudoalteromonas piscicida 2515 revealed its antibacterial potency against Vibrio anguillarum: a preliminary invitro study.
Wang F, Ghonimy A, Wang X. Wang F, et al. Antonie Van Leeuwenhoek. 2024 May 29;117(1):84. doi: 10.1007/s10482-024-01974-w. Antonie Van Leeuwenhoek. 2024. PMID: 38809302 - Identification and transcriptional control of Caulobacter crescentus genes encoding proteins containing a cold shock domain.
Lang EA, Marques MV. Lang EA, et al. J Bacteriol. 2004 Sep;186(17):5603-13. doi: 10.1128/JB.186.17.5603-5613.2004. J Bacteriol. 2004. PMID: 15317764 Free PMC article. - Heat shock response in Lactobacillus plantarum.
De Angelis M, Di Cagno R, Huet C, Crecchio C, Fox PF, Gobbetti M. De Angelis M, et al. Appl Environ Microbiol. 2004 Mar;70(3):1336-46. doi: 10.1128/AEM.70.3.1336-1346.2004. Appl Environ Microbiol. 2004. PMID: 15006751 Free PMC article.
References
- Aukrust T, Blom H. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res Int. 1992;25:253–261.
- Bae W, Phadtare S, Severinov K, Inouye M. Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol Microbiol. 1999;31:1429–1441. - PubMed
- Bernard N, Ferain T, Garmyn D, Hols P, Delcour J. Cloning of the d-lactate dehydrogenase gene from Lactobacillus delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FEBS Lett. 1991;290:61–64. - PubMed
- Bläsi U, O'Connor M, Squires C L, Dahlberg A E. Misled by sequence complementarity: does the DB-anti-DB interaction withstand scientific scrutiny? Mol Microbiol. 1999;33:439–441. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources