CaMKIIalpha 3' untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage - PubMed (original) (raw)
CaMKIIalpha 3' untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage
M S Rook et al. J Neurosci. 2000.
Abstract
The CaMKIIalpha mRNA extends into distal hippocampal dendrites, and the 3' untranslated region (3'UTR) is sufficient to mediate this localization. We labeled the 3'UTR of the CaMKIIalpha mRNA in hippocampal cultures by using a green fluorescent protein (GFP)/MS2 bacteriophage tagging system. The CaMKIIalpha 3'UTR formed discrete granules throughout the dendrites of transfected cells. The identity of the fluorescent granules was verified by in situ hybridization. Over 30 min time periods these granules redistributed without a net increase in granule number; with depolarization there is a tendency toward increased numbers of granules in the dendrites. These observations suggest that finer time resolution of granule motility might reveal changes in the motility characteristics of granules after depolarization. So that motile granules could be tracked, shorter periods of observation were required. The movements of motile granules can be categorized as oscillatory, unidirectional anterograde, or unidirectional retrograde. Colocalization of CaMKIIalpha 3'UTR granules and synapses suggested that oscillatory movements allowed the granules to sample several local synapses. Neuronal depolarization increased the number of granules in the anterograde motile pool. Based on the time frame over which the granule number increased, the translocation of granules may serve to prepare the dendrite for mounting an adequate local translation response to future stimuli. Although the resident pool of granules can respond to signals that induce local translation, the number of granules in a dendrite might reflect its activation history.
Figures
Fig. 1.
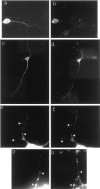
Dual constructs for GFP labeling of specific RNAs in living cells. a, The GFP fusion expression is driven by the strong CMV promotor. The MS2W82R capsid assembly-deficient RNA-binding protein is expressed as a C-terminal fusion with GFP and three copies of a consensus nuclear localization sequence. The RNA construct is expressed by the strong RSV promotor and generates a translationally competent RNA encoding the lacZ reporter gene, eight copies of the 18 bp MS2-binding site RNA hairpin, and the RNA of interest. After expression of both constructs the GFP fusion should bind as a dimer to each MS2-binding site, leading to an amplified GFP signal. b, Phase-contrast image of a transfected cell.c, A live cell expressing both constructs with the CaMKIIα 3′UTR mRNA as the RNA of interest. A higher-magnification image of the boxed region shows many small GFP-labeled granules in the dendrites. Arrows denote RNA-containing granules. d, A live cell expressing the GFP construct alone shows diffuse staining. A higher-magnification image of the_boxed region_ also shows smooth staining.
Fig. 2.
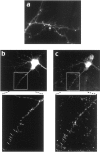
The CaMKIIα 3′UTR is necessary and sufficient to localize RNA granules to distal dendrites. GFP-labeled granules contain CaMKIIα 3′UTR RNA and components of translation. a, b, A cell transfected with the lacZ-MS2-binding site empty vector.a, β-Gal staining shows that the RNA is translationally competent and that β-gal diffuses throughout the processes. b, In situ hybridization with probes for lacZ labels punctate RNA granules only in the cell body.c, d, A cell transfected with the lacZ-MS2-binding site–CaMKIIα 3′UTR fusion construct. Figures are a composite from three separate images of the same cell. c, β-Gal staining is seen throughout the processes. d, In situ hybridization labels punctate RNA granules in the cell body and throughout the dendrites. e, f, CaMKIIα 3′UTR containing GFP-labeled granules in dendrites colocalize with lacZ-CaMKIIα 3′UTR RNA detected by _in situ_hybridization. Arrows show colocalized granules. Images were analyzed with MetaMorph software, coordinates were mapped onto images that were taken by using GFP or a rhodamine filter set, and granules were considered to colocalize when the coordinates of two objects were the same. e, GFP-labeled RNA granules.f, lacZ-CaMKIIα 3′UTR RNA detected by in situ hybridization. g, h, GFP-labeled granules colocalize with granules stained with anti-EF1α.Arrows show colocalized staining. g, GFP-labeled granules. h, EF1α staining.
Fig. 3.
Proximity of GFP-labeled CaMKIIα 3′UTR RNA granules to synapses. a, GFP-labeled CaMKIIα 3′UTR mRNA-containing granules are localized at high density in junctions (arrows) and often are seen at the surface of dendrites and at the base of spines (arrowheads). b, c, Some RNA-containing granules colocalize with synaptophysin antibody. GFP-labeled RNA granules from a KCl-treated cell are shown in b; the synaptophysin-labeled cell is shown in c. The boxed regions are shown at higher magnification below, and _arrows_show colocalized granules. A region of dendrite 3 μm long is labeled with the white bar and shows a GFP-labeled granule near five synapses labeled with synaptophysin.
Fig. 4.
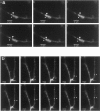
CaMKIIα 3′UTR RNA granules show both oscillatory and unidirectional motility properties.a, Time-lapse images of an oscillatory granule. Frames are sequential images taken every 20 sec. The_arrowhead_ labels an oscillatory granule, whereas an_arrow_ labels a stationary granule. b, Time-lapse of an anterograde-moving granule (arrowhead). Total distance translocated was 5.85 μm. Average velocity over 160 sec was 0.04 ± 0.01 μm/sec. The granule was docked originally at a junction, but after depolarization it moved in an anterograde direction. QuickTime movies are available for these images.
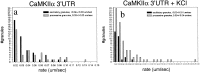
Fig. 5.
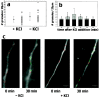
Effects of KCl on RNA granule density.a, KCl depolarization only slightly increases the granule density in dendrites. The average granule density was calculated from 35 processes from 29 different control cells and 40 processes from 24 different KCl-treated cells. b, KCl depolarization increases granule density slightly over time, but this difference does not reach statistical significance. Gray bars represent the average granule density for KCl-treated cells; black bars show the granule density for control cells. c, Time-lapse images looking at an individual process over long time periods show changes in granule distribution. Under basal conditions the images taken at 0 min and after 30 min show a redistribution of granules but no increase in granule number, whereas cells treated with KCl show both a redistribution of granules and an increase in the number of granules present. Red dots represent stationary granules that have not changed position in the 30 min recording interval, whereas_green dots_ represent new granules that have entered the region of dendrite being imaged or granules that have changed position in the dendrite during the 30 min recording interval. Granules were assigned on the basis of having a well defined circular shape and a pixel intensity above background levels in the process.
Fig. 6.
Effect of KCl depolarization on motility rates.a, The distribution of rates for CaMKIIα 3′UTR RNA-containing granules under basal conditions. Oscillatory granule rates are shown in black; unidirectional granule rates are shown in gray. Rates were calculated by measuring the change in position of a granule between two consecutive time-lapse images and dividing this distance by the time between images. b, The effect of KCl depolarization on the distribution of motility rates for CaMKIIα 3′UTR RNA-containing granules. After depolarization with KCl more granules moved unidirectionally, and more granules exhibited rates higher than 0.1 μm/sec.
Fig. 7.
KCl depolarization shifts the motility characteristics of granules from oscillatory to anterograde unidirectional. a, Motile granules fall into three directional categories: oscillatory, anterograde, and retrograde. The plots show the distance each granule traveled during a time-lapse recording at 20 sec intervals. Movements in the anterograde direction are scored as positive, and movements in the retrograde direction are scored as negative. Plots are shown for both control cells and cells treated with 10 m
m
KCl. The addition of KCl shifts the population of granules from oscillatory to anterograde.b, The distance traveled by control granules displays a gaussian histogram, whereas the histogram of the distance traveled by granules in KCl-depolarized cells is shifted toward anterograde movement.
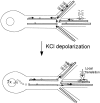
Fig. 8.
A model for the function of RNA granule motility. Under basal conditions RNA is transcribed and directed by the 3′UTR to dendrites. Within the recording interval many granules appear stationary and cluster at junctions. Some granules are motile and move in an oscillatory manner, perhaps to sample a small number of synapses. KCl depolarization activates transcription of endogenous (but not GFP-labeled) CaMKIIα mRNA and initiates CaMKIIα translation at synapses within localized RNA granules. RNA granules drawn from the oscillatory pool translocate to activated synapses where they cluster and the local translation occurs. GFP-labeled CaMKIIα 3′UTR mRNA-containing granules are shown as circles.Arrows denote moving granules; granules with_two-directional arrows_ are oscillatory. Microtubules are shown as black bars. Arrows at synapses represent activation. Tx, Transcription.
Similar articles
- An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites.
Bannai H, Fukatsu K, Mizutani A, Natsume T, Iemura S, Ikegami T, Inoue T, Mikoshiba K. Bannai H, et al. J Biol Chem. 2004 Dec 17;279(51):53427-34. doi: 10.1074/jbc.M409732200. Epub 2004 Oct 8. J Biol Chem. 2004. PMID: 15475564 - A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons.
Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. Wells DG, et al. J Neurosci. 2001 Dec 15;21(24):9541-8. doi: 10.1523/JNEUROSCI.21-24-09541.2001. J Neurosci. 2001. PMID: 11739565 Free PMC article. - mRNA at synapses, synaptic plasticity, and memory consolidation.
Steward O. Steward O. Neuron. 2002 Oct 24;36(3):338-40. doi: 10.1016/s0896-6273(02)01006-1. Neuron. 2002. PMID: 12408837 - [The mechanism for dendritic localization of alpha CaMKII mRNA].
Mori Y, Tohyama M. Mori Y, et al. Tanpakushitsu Kakusan Koso. 2001 Oct;46(13):1970-6. Tanpakushitsu Kakusan Koso. 2001. PMID: 11593749 Review. Japanese. No abstract available. - RNA transport and local protein synthesis in the dendritic compartment.
Gardiol A, Racca C, Triller A. Gardiol A, et al. Results Probl Cell Differ. 2001;34:105-28. doi: 10.1007/978-3-540-40025-7_7. Results Probl Cell Differ. 2001. PMID: 11288671 Review. No abstract available.
Cited by
- A role for dendritic translation of CaMKIIα mRNA in olfactory plasticity.
Néant-Fery M, Pérès E, Nasrallah C, Kessner M, Gribaudo S, Greer C, Didier A, Trembleau A, Caillé I. Néant-Fery M, et al. PLoS One. 2012;7(6):e40133. doi: 10.1371/journal.pone.0040133. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768241 Free PMC article. - START: A Versatile Platform for Bacterial Ligand Sensing with Programmable Performances.
Kim J, Seo M, Lim Y, Kim J. Kim J, et al. Adv Sci (Weinh). 2024 Sep;11(36):e2402029. doi: 10.1002/advs.202402029. Epub 2024 Jul 29. Adv Sci (Weinh). 2024. PMID: 39075726 Free PMC article. - In vivo monitoring of mRNA movement in Drosophila body wall muscle cells reveals the presence of myofiber domains.
van Gemert AM, van der Laan AM, Pilgram GS, Fradkin LG, Noordermeer JN, Tanke HJ, Jost CR. van Gemert AM, et al. PLoS One. 2009 Aug 17;4(8):e6663. doi: 10.1371/journal.pone.0006663. PLoS One. 2009. PMID: 19684860 Free PMC article. - Linear 2' O-Methyl RNA probes for the visualization of RNA in living cells.
Molenaar C, Marras SA, Slats JC, Truffert JC, Lemaître M, Raap AK, Dirks RW, Tanke HJ. Molenaar C, et al. Nucleic Acids Res. 2001 Sep 1;29(17):E89-9. doi: 10.1093/nar/29.17.e89. Nucleic Acids Res. 2001. PMID: 11522845 Free PMC article. - Target competition for resources under multiple search-and-capture events with stochastic resetting.
Bressloff PC. Bressloff PC. Proc Math Phys Eng Sci. 2020 Oct;476(2242):20200475. doi: 10.1098/rspa.2020.0475. Epub 2020 Oct 14. Proc Math Phys Eng Sci. 2020. PMID: 33223946 Free PMC article.
References
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. - PubMed
- Bassell GJ, Singer RH, Kosik KS. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. - PubMed
- Beach D, Salmon E, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. - PubMed
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources