Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent - PubMed (original) (raw)
Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent
A L Reysenbach et al. Appl Environ Microbiol. 2000 Sep.
Abstract
The phylogenetic diversity was determined for a microbial community obtained from an in situ growth chamber placed on a deep-sea hydrothermal vent on the Mid-Atlantic Ridge (23 degrees 22' N, 44 degrees 57' W). The chamber was deployed for 5 days, and the temperature within the chamber gradually decreased from 70 to 20 degrees C. Upon retrieval of the chamber, the DNA was extracted and the small-subunit rRNA genes (16S rDNA) were amplified by PCR using primers specific for the Archaea or Bacteria domain and cloned. Unique rDNA sequences were identified by restriction fragment length polymorphisms, and 38 different archaeal and bacterial phylotypes were identified from the 85 clones screened. The majority of the archaeal sequences were affiliated with the Thermococcales (71%) and Archaeoglobales (22%) orders. A sequence belonging to the Thermoplasmales confirms that thermoacidophiles may have escaped enrichment culturing attempts of deep-sea hydrothermal vent samples. Additional sequences that represented deeply rooted lineages in the low-temperature eurarchaeal (marine group II) and crenarchaeal clades were obtained. The majority of the bacterial sequences obtained were restricted to the Aquificales (18%), the epsilon subclass of the Proteobacteria (epsilon-Proteobacteria) (40%), and the genus Desulfurobacterium (25%). Most of the clones (28%) were confined to a monophyletic clade within the epsilon-Proteobacteria with no known close relatives. The prevalence of clones related to thermophilic microbes that use hydrogen as an electron donor and sulfur compounds (S(0), SO(4), thiosulfate) indicates the importance of hydrogen oxidation and sulfur metabolism at deep-sea hydrothermal vents. The presence of sequences that are related to sequences from hyperthermophiles, moderate thermophiles, and mesophiles suggests that the diversity obtained from this analysis may reflect the microbial succession that occurred in response to the shift in temperature and possible associated changes in the chemistry of the hydrothermal fluid.
Figures
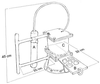
FIG. 1
Diagrammatic representation of the in situ growth chamber or vent cap. The temperature datalogger (A) and the slide mechanism (B) that opens or closes the chamber (C) are shown.
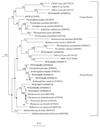
FIG. 2
Phylogenetic relationships of archaeal 16S rRNA sequences as determined by maximum-likelihood analysis. Aquifex pyrophilus was used as the outgroup. The numbers at the nodes are the bootstrap values (as percentages). The bootstrap values were less than 50% for the branch points marked with small black circles and no numerical value. Sequences from Snake Pit are marked in bold type, and the remaining sequences were obtained from the RDP (32). The scale bar represents the expected number of changes per nucleotide position.
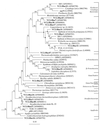
FIG. 3
Phylogenetic relationships of bacterial 16S rRNA sequences as determined by maximum-likelihood analysis. Methanococcus jannaschii was used as the outgroup. The numbers at the nodes are the bootstrap values (as percentages). The bootstrap values were less than 50% for the branch points marked with small black circles and no numerical value. Sequences from the in situ growth chamber deployed at Snake Pit are marked in bold type, and the remaining sequences were obtained from the RDP (32). The scale bar represents the expected number of changes per nucleotide position.
FIG. 4
Phylogenetic relationships of the deeply branching bacterial 16S rRNA sequences as determined by maximum-likelihood analysis. Methanococcus jannaschii was used as the outgroup. The numbers at the nodes are the bootstrap values (as percentages). Sequences from Snake Pit are marked in bold type, and the remaining sequences were obtained from the RDP (32). Cl15bon represents an unpublished sequence obtained from a second deployment of the in situ growth chamber at Snake Pit (courtesy of E. Corre). The scale bar represents the expected number of changes per nucleotide position.
Similar articles
- Microbial diversity in hydrothermal surface to subsurface environments of Suiyo Seamount, Izu-Bonin Arc, using a catheter-type in situ growth chamber.
Higashi Y, Sunamura M, Kitamura K, Nakamura K, Kurusu Y, Ishibashi J, Urabe T, Maruyama A. Higashi Y, et al. FEMS Microbiol Ecol. 2004 Mar 1;47(3):327-36. doi: 10.1016/S0168-6496(04)00004-2. FEMS Microbiol Ecol. 2004. PMID: 19712321 - Microbial diversity of a sulfide black smoker in main endeavour hydrothermal vent field, Juan de Fuca Ridge.
Zhou H, Li J, Peng X, Meng J, Wang F, Ai Y. Zhou H, et al. J Microbiol. 2009 Jun;47(3):235-47. doi: 10.1007/s12275-008-0311-z. Epub 2009 Jun 26. J Microbiol. 2009. PMID: 19557339 - Analysis of dissimilatory sulfite reductase and 16S rRNA gene fragments from deep-sea hydrothermal sites of the Suiyo Seamount, Izu-Bonin Arc, Western Pacific.
Nakagawa T, Ishibashi J, Maruyama A, Yamanaka T, Morimoto Y, Kimura H, Urabe T, Fukui M. Nakagawa T, et al. Appl Environ Microbiol. 2004 Jan;70(1):393-403. doi: 10.1128/AEM.70.1.393-403.2004. Appl Environ Microbiol. 2004. PMID: 14711668 Free PMC article. - Culturability and secondary metabolite diversity of extreme microbes: expanding contribution of deep sea and deep-sea vent microbes to natural product discovery.
Pettit RK. Pettit RK. Mar Biotechnol (NY). 2011 Feb;13(1):1-11. doi: 10.1007/s10126-010-9294-y. Epub 2010 May 1. Mar Biotechnol (NY). 2011. PMID: 20437069 Review. - Genetic techniques for the archaea.
Farkas JA, Picking JW, Santangelo TJ. Farkas JA, et al. Annu Rev Genet. 2013;47:539-61. doi: 10.1146/annurev-genet-111212-133225. Epub 2013 Sep 18. Annu Rev Genet. 2013. PMID: 24050175 Review.
Cited by
- Identification of a novel Afipia species isolated from an Indian flying fox.
Pickering BS, Tyler S, Smith G, Burton L, Li M, Dallaire A, Weingartl H. Pickering BS, et al. PLoS One. 2015 Apr 15;10(4):e0121274. doi: 10.1371/journal.pone.0121274. eCollection 2015. PLoS One. 2015. PMID: 25874801 Free PMC article. - Deep-sea hydrothermal vent bacteria related to human pathogenic Vibrio species.
Hasan NA, Grim CJ, Lipp EK, Rivera IN, Chun J, Haley BJ, Taviani E, Choi SY, Hoq M, Munk AC, Brettin TS, Bruce D, Challacombe JF, Detter JC, Han CS, Eisen JA, Huq A, Colwell RR. Hasan NA, et al. Proc Natl Acad Sci U S A. 2015 May 26;112(21):E2813-9. doi: 10.1073/pnas.1503928112. Epub 2015 May 11. Proc Natl Acad Sci U S A. 2015. PMID: 25964331 Free PMC article. - Microbial succession during the transition from active to inactive stages of deep-sea hydrothermal vent sulfide chimneys.
Hou J, Sievert SM, Wang Y, Seewald JS, Natarajan VP, Wang F, Xiao X. Hou J, et al. Microbiome. 2020 Jun 30;8(1):102. doi: 10.1186/s40168-020-00851-8. Microbiome. 2020. PMID: 32605604 Free PMC article. - Effects of dissolved sulfide, pH, and temperature on growth and survival of marine hyperthermophilic Archaea.
Lloyd KG, Edgcomb VP, Molyneaux SJ, Böer S, Wirsen CO, Atkins MS, Teske A. Lloyd KG, et al. Appl Environ Microbiol. 2005 Oct;71(10):6383-7. doi: 10.1128/AEM.71.10.6383-6387.2005. Appl Environ Microbiol. 2005. PMID: 16204562 Free PMC article. - Occurrence and diversity of yeasts in the mid-atlantic ridge hydrothermal fields near the Azores Archipelago.
Gadanho M, Sampaio JP. Gadanho M, et al. Microb Ecol. 2005 Oct;50(3):408-17. doi: 10.1007/s00248-005-0195-y. Epub 2005 Nov 24. Microb Ecol. 2005. PMID: 16328655
References
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1994.
- Blöchl E, Rachel R, Burggraf S, Hafelbradl D, Jannasch H W, Stetter K O. Pyrolobus fumarii gen. and sp. nov., represents a novel group of archaea extending the upper temperature limit for life to 113°C. Extremophiles. 1997;1:14–21. - PubMed
- Burggraf S, Jannasch H W, Nicolaus B, Stetter K O. Archaeoglobus profundus sp. nov., represents a new species within the sulfate-reducing archaebacteria. Syst Appl Microbiol. 1990;13:24–28.
- Burggraf S, Olsen G J, Stetter K O, Woese C R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:352–356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases