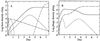Mathematical analysis of growth and interaction dynamics of streptomycetes and a bacteriophage in soil - PubMed (original) (raw)
Mathematical analysis of growth and interaction dynamics of streptomycetes and a bacteriophage in soil
N J Burroughs et al. Appl Environ Microbiol. 2000 Sep.
Abstract
We observed the infection cycle of the temperate actinophage KC301 in relation to the growth of its host Streptomyces lividans TK24 in sterile soil microcosms. Despite a large increase in phage population following germination of host spores, there was no observable impact on host population numbers as measured by direct plate counts. The only change in the host population following infection was the establishment of a small subpopulation of KC301 lysogens. The interaction of S. lividans and KC301 in soil was analyzed with a population-dynamic mathematical model to determine the underlying mechanisms of this low susceptibility to phage attack relative to aquatic environments. This analysis suggests that the soil environment is a highly significant component of the phage-host interaction, an idea consistent with earlier observations on the importance of the environment in determining host growth and phage-host dynamics. Our results demonstrate that the accepted phage-host interaction and host life cycle, as determined from agar plate studies and liquid culture, is sufficient for quantitative agreement with observations in soil, using soil-determined rates. There are four significant effects of the soil environment: (i) newly germinated spores are more susceptible to phage lysis than are hyphae of developed mycelia, (ii) substrate mycelia in mature colonies adsorb about 98% of the total phage protecting susceptible young hyphae from infection, (iii) the burst size of KC301 is large in soil (>150, 90% confidence) relative to that observed in liquid culture (120, standard error of the mean [SEM], 6), and (iv) there is no measurable impact on the host in terms of reduced growth by the phage. We hypothesize that spatial heterogeneity is the principal cause of these effects and is the primary determinant in bacterial escape of phage lysis in soil.
Figures
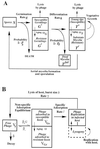
FIG. 1
(A) Schematic representation of the host dynamics. Germlings and young tips are initially susceptible to phage lysis (compartments Gs and Ts) but acquire resistance as they age at rates ω_G_ and ω_T_, respectively (resistant compartments Gr and Tr). After germination, germlings differentiate into exponentially growing substrate mycelium at rate k. Germlings are viable with probability Pg and are successful in forming colonies with probability Pd. If either probability is <1, then a drop in total propagules occurs after germination, which can alternatively be described as death rates g(1 − P g) and k(1 − P d) of spores and germlings, respectively. Germlings that die still adsorb phage nonspecifically (Tfail). In the basic model Pg is 1 and ω_T_ is large so that young tips are effectively resistant. (B) Schematic representation of the phage dynamics interacting with germlings. Interaction with substrate is similar, with T replacing G. Phage adsorbs to mycelia in a two-step process: nonspecific reversible adsorption (forward rate β+ × mycelium density G, reverse rate β−) producing phage VGs and VGr adsorbed to susceptible and resistant germlings, respectively, and specific adsorption leading to host lysis, which is modeled as an infection event (rate l) after nonspecific adsorption. This produces a replicating phage state in an infected host I. Lysis occurs at a rate γ, releasing b free-phage particles. In a model extension, lysogeny is also included as an outcome of infection. Host aging and differentiation transfer adsorbed phage between compartments following host changes. Thus, resistance can be acquired before infection by nonspecific adsorbed phage.
FIG. 2
Model simulation. (A) Host dynamics: spores (————), susceptible germlings Gs (-----), susceptible mycelia Ts (·······), resistant germlings Gr (–·–·–) and resistant mycelia Tr (———). (B) Phage dynamics: free phage Vf (———); phage adsorbed to susceptible hosts, V s = V Gs + V Ts (––––); and phage adsorbed to resistant hosts, V r = V Gr + V Tr (—-—-—). A sporulation cycle is included to demonstrate the increase in total propagule number after the cessation of exponential growth of the mycelia. Sporulation is modeled as a timed event from germination (18). The parameters in this simulation were chosen to emphasize the model properties discussed in the text: μ = 3 day−1, K = 4 × 106 CFU g−1, k = 1 day−1, P d = 0.01, b = 300, β+ = 100 day−1 CFU−1 g, β− = 1 day−1, l = 2 day−1, ω_G_ = 50 day−1, ω_T_ = 2 day−1, γ = 25 day−1, δ = 0.12 day−1, and g = 6 day−1.
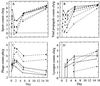
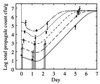
FIG. 3
Growth curves (log10) for experiment 1 (solid lines) and experiment 2 (dashed lines). (A) Spore counts. (B) Total propagules. (C) Phage counts (microcosms 2A, 2B, and 2C have less growth than microcosm 2D and are omitted for clarity). (D) Lysogen counts. For experiment 1, an average over microcosms is shown for spore and total propagules since streptomycete growth is identical across microcosms (P > 1% for microcosms 1A, 1B, and 1D). Typical error bars (95%) are shown as indicated, displaced to the right for clarity. Their skewed appearance is due to the logarithmic scale; the confidence intervals were computed based on the absolute values. Symbols (A and B): 2A, ◊; 2B, +; 2C, □; 2D, ×; 2E, ▵; 2F, ∗ with a dashed line; experiment 1 (average over microcosms), solid line. Symbols (C and D): 1A, ◊; 1B, + (experiment 1, solid line); 2D, □; 2E, ×; 2F, ▵ (experiment 2, dashed line). Data are reprinted from Gene Transfers and Environment (25) with permission from the publisher.
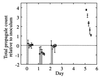
FIG. 4
Plot of total propagule growth relative to the inoculum against time on a log10 scale. Microcosms 2C to 2F and an average over microcosms for experiment 1 (adjusted for the difference in extraction efficiency) are shown. Symbols: 2C, ◊; 2D, +; 2E, □; 2F, ×; experiment 1, ▵.
FIG. 5
Fit of basic model to total propagule data of experiment 2, microcosms 2B to 2F. The data are time displaced for clarity. Error bars are 95%. The parameters are given in Table 3. Note the uniformity in the growth over the first 3 days, which reproduces the scaling behavior of Fig. 4. Symbols: 2B, ◊ (———); 2C, + (————); 2D, □ (–––––); 2E, × (-----); 2F, ▵, (—-—-—).
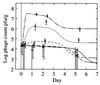
FIG. 6
Fit of model to phage data for microcosms 2B to 2F. Symbols: 2B, ◊ (———); 2C, + (————); 2D, □ (––––); 2E, × (------); 2F, ▵ (—-—-—).
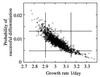
FIG. 7
Correlation between parameters Pd, the probability of a germling differentiating successfully, and μ, the mycelium growth rate. The results are based on 2,000 Monte Carlo simulations. The 90% confidence intervals of the projected distributions are shown. The joint 90% confidence interval calculated from the SS lies inside these separate confidence intervals.
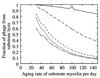
FIG. 8
The proportion of phage that originated from lysis of substrate mycelia as a function of the aging rate ω_T_ in units per day. Microcosms 2B to 2F are shown in decreasing sequence, i.e., 2B (———), 2C (————), 2D (––––), 2E (·······), and 2F (-—-—-). Germlings acquired resistance at a rate ω_G_ that remained approximately constant at 1.3 day−1. In microcosm 2B, the phage growth was so low that changes in numerical accuracy of simulation are apparent at a ω_T_ of 100 day−1 compared to surrounding points.
Similar articles
- Gene transfer between streptomycetes in soil.
Wellington EM, Cresswell N, Herron PR. Wellington EM, et al. Gene. 1992 Jun 15;115(1-2):193-8. doi: 10.1016/0378-1119(92)90559-8. Gene. 1992. PMID: 1612437 - Exclusion of induced bacteriophage from cells of a lysogenic Bacillus megaterium committed to sporulation.
Hendry GS, Fitz-James PC. Hendry GS, et al. J Bacteriol. 1974 Apr;118(1):295-303. doi: 10.1128/jb.118.1.295-303.1974. J Bacteriol. 1974. PMID: 4206875 Free PMC article. - Use of polyvalent phage for reduction of streptomycetes on soil dilution plates.
Kurtböke DI, Chen CF, Williams ST. Kurtböke DI, et al. J Appl Bacteriol. 1992 Feb;72(2):103-11. doi: 10.1111/j.1365-2672.1992.tb01810.x. J Appl Bacteriol. 1992. PMID: 1556035 - Bacterial sensitivity to bacteriophage in the aquatic environment.
Day M. Day M. Sci Prog. 2004;87(Pt 3):179-91. doi: 10.3184/003685004783238526. Sci Prog. 2004. PMID: 15884658 Free PMC article. Review. - Molecular Basis of Lysis-Lysogeny Decisions in Gram-Positive Phages.
Brady A, Felipe-Ruiz A, Gallego Del Sol F, Marina A, Quiles-Puchalt N, Penadés JR. Brady A, et al. Annu Rev Microbiol. 2021 Oct 8;75:563-581. doi: 10.1146/annurev-micro-033121-020757. Epub 2021 Aug 3. Annu Rev Microbiol. 2021. PMID: 34343015 Review.
Cited by
- Phage-host interaction: an ecological perspective.
Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brüssow H. Chibani-Chennoufi S, et al. J Bacteriol. 2004 Jun;186(12):3677-86. doi: 10.1128/JB.186.12.3677-3686.2004. J Bacteriol. 2004. PMID: 15175280 Free PMC article. Review. No abstract available. - In situ monitoring of streptothricin production by Streptomyces rochei F20 in soil and rhizosphere.
Anukool U, Gaze WH, Wellington EM. Anukool U, et al. Appl Environ Microbiol. 2004 Sep;70(9):5222-8. doi: 10.1128/AEM.70.9.5222-5228.2004. Appl Environ Microbiol. 2004. PMID: 15345403 Free PMC article. - Horizontal gene exchange in environmental microbiota.
Aminov RI. Aminov RI. Front Microbiol. 2011 Jul 26;2:158. doi: 10.3389/fmicb.2011.00158. eCollection 2011. Front Microbiol. 2011. PMID: 21845185 Free PMC article. - Dynamics of Bacterial and Viral Communities in Paddy Soil with Irrigation and Urea Application.
Li Y, Sun H, Yang W, Chen G, Xu H. Li Y, et al. Viruses. 2019 Apr 16;11(4):347. doi: 10.3390/v11040347. Viruses. 2019. PMID: 31014039 Free PMC article. - Horizontal gene transfer in the phytosphere.
Van Elsas JD, Turner S, Bailey MJ. Van Elsas JD, et al. New Phytol. 2003 Mar;157(3):525-537. doi: 10.1046/j.1469-8137.2003.00697.x. New Phytol. 2003. PMID: 33873398 Review.
References
- Adams M H. Bacteriophages. New York, N.Y: Interscience Publishers, Inc.; 1959.
- Allan E J, Prosser J I. A kinetic study of the colony growth of Streptomyces coelicolor A3(2) and J802 on solid medium. J Gen Microbiol. 1985;131:2521–2532.
- Allan E J, Prosser J I. Colony growth of Streptomyces coelicolor A3(2) under conditions of continuous nutrient supply. FEMS Microbiol Lett. 1987;43:139–142.
- Bader F G. Sterilization: prevention of contamination. In: Demain A L, Solomon N A, editors. Manual of industrial microbiology and biotechnology. Washington, D.C.: American Society for Microbiology; 1986. pp. 345–362.
- Beale E M L. Confidence regions in non-linear estimation. J R Stat Soc. 1960;22B:41–88.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources