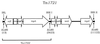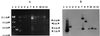Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tet A - PubMed (original) (raw)
Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tet A
G Rhodes et al. Appl Environ Microbiol. 2000 Sep.
Abstract
Oxytetracycline-resistant (OT(r)) mesophilic aeromonads were recovered from untreated hospital effluent (72 isolates) and from fish farm hatchery tanks (91 isolates) at sites within the English Lake District, Cumbria, England. The transfer of OT(r) plasmids from these isolates was investigated. Using Escherichia coli J53-1 as a recipient, 11 isolates from the hospital site and 6 isolates from the fish farm site transferred OT(r) plasmids (designated pFBAOT1 to 17). Original isolates were identified using fatty acid methyl ester and fluorescent amplified fragment length polymorphism comparisons as either Aeromonas hydrophila HG3 (eight isolates), A. veronii b.v. sobria HG8 (six isolates), and A. caviae HGB5 (one isolate). One isolate remained unidentified, and one could not be assigned a taxonomic designation beyond the genus level. Plasmids pFBAOT1 to -17 were screened for the presence of the tetracycline resistance determinants Tet A to E and Tet G. Only determinant Tet A (10 plasmids) was detected in these plasmids, with 7 tet gene determinants remaining unclassified. In all cases, Tet A was located on a 5.5-kb EcoRI restriction fragment. Hybridization with inc-rep probes N, P, Q, W, and U showed pFBAOT3, -4, -5, -6, -7, -9, and -11, from the hospital environment, to be IncU plasmids. Further, restriction fragment length polymorphism (RFLP) analyses and DNA probing demonstrated that pFBAOT plasmids were closely related to IncU OT(r) plasmids pASOT, pASOT2, pASOT3, pRAS1 (originally isolated from A. salmonicida strains from fish farms in Scotland and Norway, respectively), and pIE420 (isolated from a German hospital E. coli strain). In addition, DNA analyses demonstrated that plasmids pRAS1 and pIE420 had identical RFLP profiles and that all fragments hybridized to each other. The presence of tetracycline resistance transposon Tn1721 in its entirety or in a truncated form in these plasmids was demonstrated. These results provided direct evidence that related tetracycline resistance-encoding plasmids have disseminated between different Aeromonas species and E. coli and between the human and aquaculture environments in distinct geographical locations. Collectively, these findings provide evidence to support the hypothesis that the aquaculture and human compartments of the environment behave as a single interactive compartment.
Figures
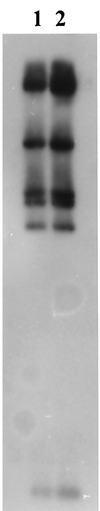
FIG. 1
(A) RFLP profiles (generated by _Eco_RI digestion) of IncU OTr plasmids from aeromonads and corresponding Southern hybridizations with determinant Tet A and pASOT plasmid. Lanes: 1 and 11, λ DNA markers digested with _Hin_dIII and a 1-kb ladder, respectively; 2, pIE420; 3, pASOT; 4, pFBAOT3; 5, pFBAOT4; 6, pFBAOT5; 7, pFBAOT6; 8, pFBAOT7; 9, pFBAOT9; 10, pFBAOT11. (B) Hybridization of the Tet A determinant probe to plasmids. All plasmids harbored Tet A on a 5.5-kb fragment. (C) Hybridization of plasmid pASOT to IncU plasmids.
FIG. 2
Comparison of plasmids pRAS1 and pIE420 by hybridization of radiolabeled plasmid pRAS1 to both plasmids. Lanes: 1, pRAS1; 2, pIE420.
FIG. 3
Physical and genetic structure of transposon Tn_1721_ (length, 11,139 bp). The 38-bp inverted repeats IRL, IRR-I, and IRR-II are highlighted. The designation, position, and direction of transcription of genes are shown by the arrowed boxes. The region that comprises the minor transposon Tn_1722_ is highlighted, along with the location of _Eco_RI restriction sites. This diagram was adapted from Schnabel and Jones (39) and Tsuda et al. (45).
FIG. 4
(A) _Eco_RI-generated RFLP patterns for IncU OTr plasmids. The gel was electrophoresed for an extended period in order to separate the fragments in the 5- to 7-kb range. Lanes: 1 and 12, λ DNA markers digested with _Hin_dIII and a 1-kb ladder, respectively; 2, pMT1286; 3, pASOT; 4, pIE420; 5, pFBAOT3; 6, pFBAOT4; 7, pFBAOT5; 8, pFBAOT6; 9, pFBAOT7; 10, pFBAOT9; 11, pFBAOT11; 12, 1-kb marker DNA. (B) Southern hybridization of the MCP probe to the DNA in panel A.
Similar articles
- Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems.
Jacobs L, Chenia HY. Jacobs L, et al. Int J Food Microbiol. 2007 Mar 20;114(3):295-306. doi: 10.1016/j.ijfoodmicro.2006.09.030. Epub 2006 Dec 14. Int J Food Microbiol. 2007. PMID: 17173998 - Characterization of oxytetracycline-resistant heterotrophic bacteria originating from hospital and freshwater fishfarm environments in England and Ireland.
Huys G, Rhodes G, McGann P, Denys R, Pickup R, Hiney M, Smith P, Swings J. Huys G, et al. Syst Appl Microbiol. 2000 Dec;23(4):599-606. doi: 10.1016/s0723-2020(00)80036-3. Syst Appl Microbiol. 2000. PMID: 11249032 - Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida.
Adams CA, Austin B, Meaden PG, McIntosh D. Adams CA, et al. Appl Environ Microbiol. 1998 Nov;64(11):4194-201. doi: 10.1128/AEM.64.11.4194-4201.1998. Appl Environ Microbiol. 1998. PMID: 9797265 Free PMC article. - Comparison of the antimicrobial tolerance of oxytetracycline-resistant heterotrophic bacteria isolated from hospital sewage and freshwater fishfarm water in Belgium.
Huys G, Gevers D, Temmerman R, Cnockaert M, Denys R, Rhodes G, Pickup R, McGann P, Hiney M, Smith P, Swings J. Huys G, et al. Syst Appl Microbiol. 2001 Apr;24(1):122-30. doi: 10.1078/0723-2020-00008. Syst Appl Microbiol. 2001. PMID: 11403391 - Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment.
Schmidt AS, Bruun MS, Dalsgaard I, Larsen JL. Schmidt AS, et al. Appl Environ Microbiol. 2001 Dec;67(12):5675-82. doi: 10.1128/AEM.67.12.5675-5682.2001. Appl Environ Microbiol. 2001. PMID: 11722922 Free PMC article.
Cited by
- Prevalence, Virulence Gene Distribution and Alarming the Multidrug Resistance of Aeromonas hydrophila Associated with Disease Outbreaks in Freshwater Aquaculture.
Nhinh DT, Le DV, Van KV, Huong Giang NT, Dang LT, Hoai TD. Nhinh DT, et al. Antibiotics (Basel). 2021 May 4;10(5):532. doi: 10.3390/antibiotics10050532. Antibiotics (Basel). 2021. PMID: 34064504 Free PMC article. - Call of the wild: antibiotic resistance genes in natural environments.
Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Allen HK, et al. Nat Rev Microbiol. 2010 Apr;8(4):251-9. doi: 10.1038/nrmicro2312. Epub 2010 Mar 1. Nat Rev Microbiol. 2010. PMID: 20190823 Review. - Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences.
Smet A, Van Nieuwerburgh F, Vandekerckhove TT, Martel A, Deforce D, Butaye P, Haesebrouck F. Smet A, et al. PLoS One. 2010 Jun 18;5(6):e11202. doi: 10.1371/journal.pone.0011202. PLoS One. 2010. PMID: 20585456 Free PMC article. - Antibiotic Abuse in Ornamental Fish: An Overlooked Reservoir for Antibiotic Resistance.
Au-Yeung C, Tsui YL, Choi MH, Chan KW, Wong SN, Ling YK, Lam CM, Lam KL, Mo WY. Au-Yeung C, et al. Microorganisms. 2025 Apr 18;13(4):937. doi: 10.3390/microorganisms13040937. Microorganisms. 2025. PMID: 40284775 Free PMC article. Review. - Detection of Antibiotic Resistance Genes in Source and Drinking Water Samples from a First Nations Community in Canada.
Fernando DM, Tun HM, Poole J, Patidar R, Li R, Mi R, Amarawansha GEA, Fernando WGD, Khafipour E, Farenhorst A, Kumar A. Fernando DM, et al. Appl Environ Microbiol. 2016 Jul 15;82(15):4767-4775. doi: 10.1128/AEM.00798-16. Print 2016 Aug 1. Appl Environ Microbiol. 2016. PMID: 27235436 Free PMC article.
References
- Allmeier H, Cresnar B, Greck M, Schmitt R. Complete nucleotide sequence of Tn1721: gene organisation and a novel gene product with features of a chemotaxis protein. Gene. 1992;111:11–20. - PubMed
- Aoki T, Kitao T, Iemura N, Mitoma Y, Nomura T. The susceptibility of Aeromonas salmonicida strains isolated in cultured and wild salmonids to various chemotherapeutants. Bull Jpn Soc Sci Fish. 1983;49:17–22.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources