Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism - PubMed (original) (raw)
Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism
B Prakash et al. EMBO J. 2000.
Abstract
The interferon-gamma-induced guanylate-binding protein 1 (GBP1) belongs to a special class of large GTP- binding proteins of 60-100 kDa with unique characteristics. Here we present the structure of human GBP1 in complex with the non-hydrolysable GTP analogue GppNHp. Basic features of guanine nucleotide binding, such as the P-loop orientation and the Mg(2+) co-ordination, are analogous to those of Ras-related and heterotrimeric GTP-binding proteins. However, the glycosidic bond and thus the orientation of the guanine base and its interaction with the protein are very different. Furthermore, two unique regions around the base and the phosphate-binding areas, the guanine and the phosphate caps, respectively, give the nucleotide-binding site a unique appearance not found in the canonical GTP-binding proteins. The phosphate cap, which constitutes the region analogous to switch I, completely shields the phosphate-binding site from solvent such that a potential GTPase-activating protein cannot approach. This has consequences for the GTPase mechanism of hGBP1 and possibly of other large GTP-binding proteins.
Figures
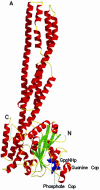
Fig. 1. Overall structure of hGBP1⋅GppNHp and comparison with nucleotide-free protein. (A) Ribbon diagram of the structure, with GppNHp as a blue stick model and Mg2+ as a yellow sphere. (B) Stereo view of a superposition of the apo (red) and GppNHp-bound (green) structures of hGBP1, as a worm plot, highlighting important regions. The regions not visible in the apo or nucleotide-bound structures are shown by a dotted line. Coloured balls [compare with (C)] mark the beginning and end of regions invisible in the apo structure. (C) R.m.s. deviation plot for the main chain (Cα) atoms of the apo and GppNHp structures, highlighting regions that show the highest deviations and/or those that were undefined in the apo structure.
Fig. 1. Overall structure of hGBP1⋅GppNHp and comparison with nucleotide-free protein. (A) Ribbon diagram of the structure, with GppNHp as a blue stick model and Mg2+ as a yellow sphere. (B) Stereo view of a superposition of the apo (red) and GppNHp-bound (green) structures of hGBP1, as a worm plot, highlighting important regions. The regions not visible in the apo or nucleotide-bound structures are shown by a dotted line. Coloured balls [compare with (C)] mark the beginning and end of regions invisible in the apo structure. (C) R.m.s. deviation plot for the main chain (Cα) atoms of the apo and GppNHp structures, highlighting regions that show the highest deviations and/or those that were undefined in the apo structure.
Fig. 1. Overall structure of hGBP1⋅GppNHp and comparison with nucleotide-free protein. (A) Ribbon diagram of the structure, with GppNHp as a blue stick model and Mg2+ as a yellow sphere. (B) Stereo view of a superposition of the apo (red) and GppNHp-bound (green) structures of hGBP1, as a worm plot, highlighting important regions. The regions not visible in the apo or nucleotide-bound structures are shown by a dotted line. Coloured balls [compare with (C)] mark the beginning and end of regions invisible in the apo structure. (C) R.m.s. deviation plot for the main chain (Cα) atoms of the apo and GppNHp structures, highlighting regions that show the highest deviations and/or those that were undefined in the apo structure.
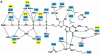
Fig. 2. Binding of nucleotide to hGBP1. (A) Schematic diagram of the interactions of GppNHp, Mg2+ and selected water molecules with the protein; dashed lines indicate contacts with distances <3.4 Å, where NHmc and COmc indicate main chain interactions. Square boxes are residues from GBP1, whereas analogous residues from Ras with similar function are in oval boxes. (B) Superposition of GppNHp bound to hGBP1 (grey) with that from the Ras⋅GppNHp structure (yellow) (Pai et al., 1990). The yellow sphere indicates the Mg2+ ion in both structures. (C) Interactions of the guanine base with selected residues and parts of the corresponding polypeptide chain of hGBP1 (in green), compared with the position of the NKCD motif in the Ras polypeptide chain (in blue), with the position of residues indicated by red balls. Red dotted lines show the interactions of the RD guanine base-binding motif.
Fig. 2. Binding of nucleotide to hGBP1. (A) Schematic diagram of the interactions of GppNHp, Mg2+ and selected water molecules with the protein; dashed lines indicate contacts with distances <3.4 Å, where NHmc and COmc indicate main chain interactions. Square boxes are residues from GBP1, whereas analogous residues from Ras with similar function are in oval boxes. (B) Superposition of GppNHp bound to hGBP1 (grey) with that from the Ras⋅GppNHp structure (yellow) (Pai et al., 1990). The yellow sphere indicates the Mg2+ ion in both structures. (C) Interactions of the guanine base with selected residues and parts of the corresponding polypeptide chain of hGBP1 (in green), compared with the position of the NKCD motif in the Ras polypeptide chain (in blue), with the position of residues indicated by red balls. Red dotted lines show the interactions of the RD guanine base-binding motif.
Fig. 3. Crystal contacts of hGBP1⋅GppNHp. (A) The head-to-tail dimer A, which buries 2890 Å2 of surface area, where a lip from the LG domain is close to the helical domain in helices α10 and the long helix α12. The nucleotide is shown in ball and stick representation with the yellow sphere representing the Mg2+ ion. (B) The head-to-head dimer B with 2140 Å2 of buried surface, using a similar colour code.
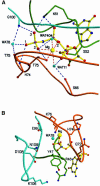
Fig. 4. The phosphate-binding region and implications for GTP hydrolysis. (A) Interactions of the phosphate oxygens and Mg2+ with the P-loop (green), the switch I/phosphate cap (brown) and the switch II region (maroon). Wat8 is in a homologous position to the nucleophilic water found in other structures of GTP-binding proteins. In contrast to those, there are three main chain NH interactions of the protein with the γ-phosphate. (B) Potential catalytic residues around the active site that could modify the rate of the GTPase reaction in an oligomerization-dependent manner, without directly participating in catalysis. (C and D) van der Waals surface representation of the region of the active site of Ras (C) and hGBP1 (D) in the GppNHp-bound state, the surface being coloured according to the electrostatic potential, as calculated with GRASP (Nicholls et al., 1991). In hGBP1, only the base is open to the solvent.
Fig. 4. The phosphate-binding region and implications for GTP hydrolysis. (A) Interactions of the phosphate oxygens and Mg2+ with the P-loop (green), the switch I/phosphate cap (brown) and the switch II region (maroon). Wat8 is in a homologous position to the nucleophilic water found in other structures of GTP-binding proteins. In contrast to those, there are three main chain NH interactions of the protein with the γ-phosphate. (B) Potential catalytic residues around the active site that could modify the rate of the GTPase reaction in an oligomerization-dependent manner, without directly participating in catalysis. (C and D) van der Waals surface representation of the region of the active site of Ras (C) and hGBP1 (D) in the GppNHp-bound state, the surface being coloured according to the electrostatic potential, as calculated with GRASP (Nicholls et al., 1991). In hGBP1, only the base is open to the solvent.
Similar articles
- How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. Ghosh A, et al. Nature. 2006 Mar 2;440(7080):101-4. doi: 10.1038/nature04510. Nature. 2006. PMID: 16511497 - The guanine cap of human guanylate-binding protein 1 is responsible for dimerization and self-activation of GTP hydrolysis.
Wehner M, Kunzelmann S, Herrmann C. Wehner M, et al. FEBS J. 2012 Jan;279(2):203-10. doi: 10.1111/j.1742-4658.2011.08415.x. Epub 2011 Nov 30. FEBS J. 2012. PMID: 22059445 - Nucleotide binding and self-stimulated GTPase activity of human guanylate-binding protein 1 (hGBP1).
Kunzelmann S, Praefcke GJ, Herrmann C. Kunzelmann S, et al. Methods Enzymol. 2005;404:512-27. doi: 10.1016/S0076-6879(05)04045-0. Methods Enzymol. 2005. PMID: 16413296 - The guanine nucleotide-binding switch in three dimensions.
Vetter IR, Wittinghofer A. Vetter IR, et al. Science. 2001 Nov 9;294(5545):1299-304. doi: 10.1126/science.1062023. Science. 2001. PMID: 11701921 Review. - GEFs, GAPs, GDIs and effectors: taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins.
Geyer M, Wittinghofer A. Geyer M, et al. Curr Opin Struct Biol. 1997 Dec;7(6):786-92. doi: 10.1016/s0959-440x(97)80147-9. Curr Opin Struct Biol. 1997. PMID: 9434896 Review.
Cited by
- Invited review: Mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily.
Daumke O, Praefcke GJ. Daumke O, et al. Biopolymers. 2016 Aug;105(8):580-93. doi: 10.1002/bip.22855. Biopolymers. 2016. PMID: 27062152 Free PMC article. Review. - The Large GTPase Guanylate-Binding Protein-1 (GBP-1) Promotes Mitochondrial Fission in Glioblastoma.
Kalb RC, Nyabuto GO, Morran MP, Maity S, Justinger JS, Nestor-Kalinoski AL, Vestal DJ. Kalb RC, et al. Int J Mol Sci. 2024 Oct 19;25(20):11236. doi: 10.3390/ijms252011236. Int J Mol Sci. 2024. PMID: 39457021 Free PMC article. - Molecular engineering of myosin.
Manstein DJ. Manstein DJ. Philos Trans R Soc Lond B Biol Sci. 2004 Dec 29;359(1452):1907-12. doi: 10.1098/rstb.2004.1565. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15647166 Free PMC article. Review. - Detection of Cytosolic Shigella flexneri via a C-Terminal Triple-Arginine Motif of GBP1 Inhibits Actin-Based Motility.
Piro AS, Hernandez D, Luoma S, Feeley EM, Finethy R, Yirga A, Frickel EM, Lesser CF, Coers J. Piro AS, et al. mBio. 2017 Dec 12;8(6):e01979-17. doi: 10.1128/mBio.01979-17. mBio. 2017. PMID: 29233899 Free PMC article. - The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines.
Guenzi E, Töpolt K, Cornali E, Lubeseder-Martellato C, Jörg A, Matzen K, Zietz C, Kremmer E, Nappi F, Schwemmle M, Hohenadl C, Barillari G, Tschachler E, Monini P, Ensoli B, Stürzl M. Guenzi E, et al. EMBO J. 2001 Oct 15;20(20):5568-77. doi: 10.1093/emboj/20.20.5568. EMBO J. 2001. PMID: 11598000 Free PMC article.
References
- Ahmadian M.R., Stege,P., Scheffzek,K. and Wittinghofer,A. (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nature Struct. Biol., 4, 686–689. - PubMed
- Anderson S.L., Carton,J.M., Lou,J., Xing,L. and Rubin,B.Y. (1999) Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalo myocarditis virus. Virology, 256, 8–14. - PubMed
- Barylko B., Binns,D., Lin,K.M., Atkinson,M.A., Jameson,D.M., Yin,H.L. and Albanesi,J.P. (1998) Synergistic activation of dynamin GTPase by Grb2 and phosphoinositides. J. Biol. Chem., 273, 3791–3797. - PubMed
- Berchtold H., Reshetnikova,L., Reiser,C.O., Schirmer,N.K., Sprinzl,M. and Hilgenfeld,R. (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature, 365, 126–132. - PubMed
- Bollag G. and McCormick,F. (1991) Differential regulation of RasGAP and neurofibromatosis gene product activities. Nature, 351, 576–579. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous