Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun - PubMed (original) (raw)
Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun
F Macián et al. EMBO J. 2000.
Abstract
Cooperation between nuclear factor of activated T cells (NFAT) and AP-1 (Fos-Jun) proteins on composite NFAT-AP-1 DNA elements constitutes a powerful mechanism for signal integration of the calcium and protein kinase C/Ras pathways in the regulation of gene expression. Here we report that NFAT can induce expression of certain genes in T cells without the need for cooperative recruitment of Fos and Jun. Using NFAT1 mutant proteins that are unable to interact with Fos-Jun dimers but are unaffected in DNA binding or transcriptional activity, we show that expression of interleukin (IL)-2, granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-3, IL-4, MIP1alpha and Fas ligand mRNAs is absolutely dependent on cooperation between NFAT and Fos-Jun; in contrast, NFAT induces tumor necrosis factor alpha (TNFalpha) mRNA and IL-13 promoter activity without any necessity to recruit Fos and Jun. Furthermore, we show that NFAT-Fos-Jun cooperation is also essential to elicit the NFAT-dependent program of activation-induced cell death. Our results support the hypothesis that even in a single cell type, NFAT activation can evoke two distinct biological programs of gene expression, dependent or independent of NFAT-AP-1 cooperation.
Figures
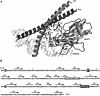
Fig. 1. Residues involved in NFAT1–Fos–Jun interaction. (A) Close-up view of the NFAT1–Fos–Jun–DNA complex (Chen et al., 1998). Ribbon diagram made using Rasmol. Side chains of NFAT1 involved in NFAT1–Fos, NFAT1–Jun and NFAT1–DNA interactions that were mutated in this study are shown. (B) Amino acid sequence and secondary structure of the NFAT1 DBD. Residues that contact Fos are shown in open boxes while residues involved in Jun contacts are shown in white inside black boxes. The residues mutated in this study are indicated by asterisks.
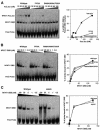
Fig. 2. The R468E and T535R mutations in the NFAT1 DBD strongly influence the interaction of NFAT1 with Fos and Jun. (A) Jun homodimers. (B) Fos–Jun heterodimers. EMSAs were performed with the murine IL-2 promoter ARRE2 probe and wild-type (left panels), R468E (middle panels) or T535R (right panels) recombinant NFAT1 DBDs. Right panels show results of densitometric quantification.
Fig. 3. Mutations that impair the ability of NFAT to interact with Fos and Jun have no effect on its DNA-binding activity. EMSAs were performed using the murine IL-2 promoter ARRE2 probe and recombinant NFAT1 DBD proteins. Right panels show results of densitometric quantification. (A) Ability of wild-type NFAT1 DBD and two mutant DBDs, F475A and R468A/I469A/T535G, to bind cooperatively with Fos and Jun to DNA. (B) The same probe was used to measure the DNA-binding activity of wild-type NFAT1 DBD and the two mutants F475A and R468A/I469A/T535G. (C) Effect of the H427R mutation on NFAT1 DNA-binding activity.
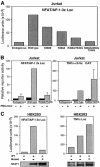
Fig. 4. Mutations that impair the interaction of NFAT1 with AP-1 also selectively impair its transcriptional activity on a composite NFAT–AP-1 site. (A) Jurkat cells were cotransfected with the NFAT/AP-1 3×-luciferase reporter plasmid and expression plasmids encoding wild-type NFAT1 or different AP-1 interaction mutants. Activity was measured after 8 h of stimulation with PMA and ionomycin. (B) Jurkat cells were cotransfected with expression plasmids encoding wild-type NFAT1 or R468A/T535G mutant NFAT1 and either a luciferase reporter plasmid driven by three copies of the IL-2 promoter ARRE-2 site (NFAT/AP-1 3×Luc; left panel) or a CAT reporter plasmid carrying five copies of the κ3 element of the TNFα promoter (κ3-5×CAT; right panel). Where indicated, the cells were stimulated with PMA and ionomycin for 8 h. Values are mean + SE of three independent experiments. A slight (2-fold) enhancement was seen with the mutant relative to the wild-type protein in the κ3-5×CAT reporter experiments, but this did not reach significant levels. (C) HEK293 cells were cotransfected with expression plasmids encoding wild-type NFAT1 or R468A/I469A/T535G mutant NFAT1 and with the NFAT/AP-1 3× (left panel) or TNFα promoter (right panel) luciferase reporter plasmids. Reporter activity was measured after 8 h of stimulation with PMA and ionomycin. Lower panels show western blots using an anti-NFAT1 antibody to detect expression of wild-type and mutant NFAT1.
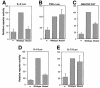
Fig. 5. Different NFAT-dependent cytokine gene promoters show differing requirements for cooperation of NFAT with AP-1. Jurkat cells were cotransfected with expression plasmids encoding wild-type NFAT1 or R468A/I469A/T535G mutant NFAT1 and luciferase reporter plasmids bearing the promoter regions of the (A) IL-2, (B) TNFα, (C) GM-CSF, (D) IL-4 and (E) IL-13 genes. Reporter activity was measured in cells activated with PMA and ionomycin for 8 h. (–) indicates the level of endogenous reporter activity in cells transfected with a control empty plasmid. Results are mean + SE of three independent experiments.
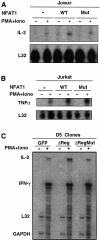
Fig. 6. AP-1-dependent and -independent expression patterns of endogenous NFAT target genes. (A) Jurkat cells were cotransfected with expression plasmids encoding murine CD4 and wild-type or R468A/I469A/T535G mutant NFAT1 (1 µg of each expression plasmid/106 cells). After 24 h, transfected cells were selected with CD4–Dynabeads and left unstimulated or stimulated with PMA and ionomycin for 4 h. Levels of expression of IL-2 (A) and TNFα (B) were determined by RNase protection assay. Levels of protected L32 transcripts indicate mRNA loading in each lane. (C) RNA from D5 clones stably expressing GFP, wild-type NFAT1ΔReg or mutant NFAT1ΔReg[R468A/T535G] were analyzed by RNase protection assay using a multi-set probe that allowed detection of IL-2 and IFNγ mRNAs. Where indicated, cells were stimulated with PMA and ionomycin for 4 h. Levels of L32 and GAPDH mRNAs show similar loading in all lanes.
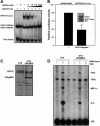
Fig. 7. The NFAT1ΔReg[R468A/T535G] mutant protein shows selective dominant-negative activity on composite NFAT–AP-1 sites. (A) The ability of the mutant R468A/T535G NFAT1 DBD (DBDMut) to compete with wild-type NFAT1 DBD (DBDWT) for binding to an NFAT–AP-1 composite site was evaluated by EMSA using the murine ARRE-2 probe. Fixed concentrations of Jun (20 nM), Fos (20 nM) and recombinant DBDWT (4 nM) were used in all binding reactions and competition by increasing amounts of DBDMut was monitored by the decrease in intensity of the band corresponding to the wild-type Fos–Jun–NFAT1 DBD complex (NFAT1:Fos:Jun). The migration positions of DNA–protein complexes containing NFAT1 DBDWT and DBDMut are indicated. (B) Jurkat cells were transiently transfected with the NFAT/AP-1 3×luciferase reporter plasmid with or without an expression plasmid encoding mutant NFAT1ΔReg[R468A/T535G], and luciferase activity was determined in cells activated with PMA and ionomycin for 8 h. A western blot showing the levels of expression of full-length endogenous NFAT1 and the mutant NFAT1ΔReg protein in transfected cells is shown (inset). (C) A D10 clone stably expressing NFAT1ΔReg[R468A/T535G] was generated by retroviral infection of D10 cells and subsequent selection. The level of protein expression achieved in this clone was determined by western blotting using an anti-NFAT1 antibody. (D) The pattern of cytokine expression of a D10 clone that expressed R468A/T535G NFAT1 ΔReg was analyzed by RNase protection assay using a multi-set cytokine probe. Where indicated, cells were stimulated with PMA and ionomycin for 4 h in the presence or absence of CsA. Levels of protected L32 transcripts show that loading was similar in all lanes.
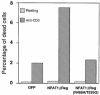
Fig. 8. Activation-induced cell death of the D5 T cell clone is enhanced by overexpression of wild-type NFAT1ΔReg but not the AP-1-interaction mutant NFAT1ΔReg[R468A/T535G]. D5 clones that expressed GFP, wild-type NFAT1ΔReg or the AP-1-interaction mutant NFAT1ΔReg[R468A/T535G] were either left unstimulated or stimulated with plate-bound anti-CD3 (1 µg/ml). After 48 h, cells were stained with propidium iodide and the proportion of dead cells was determined.
Similar articles
- Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA.
Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Chen L, et al. Nature. 1998 Mar 5;392(6671):42-8. doi: 10.1038/32100. Nature. 1998. PMID: 9510247 - Comparative analysis of NFAT (nuclear factor of activated T cells) complex in human T and B lymphocytes.
Yaseen NR, Maizel AL, Wang F, Sharma S. Yaseen NR, et al. J Biol Chem. 1993 Jul 5;268(19):14285-93. J Biol Chem. 1993. PMID: 8314792 - Control of the orientation of Fos-Jun binding and the transcriptional cooperativity of Fos-Jun-NFAT1 complexes.
Ramirez-Carrozzi VR, Kerppola TK. Ramirez-Carrozzi VR, et al. J Biol Chem. 2001 Jun 15;276(24):21797-808. doi: 10.1074/jbc.M101494200. Epub 2001 Mar 19. J Biol Chem. 2001. PMID: 11259418 - Partners in transcription: NFAT and AP-1.
Macián F, López-Rodríguez C, Rao A. Macián F, et al. Oncogene. 2001 Apr 30;20(19):2476-89. doi: 10.1038/sj.onc.1204386. Oncogene. 2001. PMID: 11402342 Review. - Transcription factors of the NFAT family: regulation and function.
Rao A, Luo C, Hogan PG. Rao A, et al. Annu Rev Immunol. 1997;15:707-47. doi: 10.1146/annurev.immunol.15.1.707. Annu Rev Immunol. 1997. PMID: 9143705 Review.
Cited by
- Attenuation of microglial activation in a mouse model of Alzheimer's disease via NFAT inhibition.
Rojanathammanee L, Floden AM, Manocha GD, Combs CK. Rojanathammanee L, et al. J Neuroinflammation. 2015 Mar 4;12:42. doi: 10.1186/s12974-015-0255-2. J Neuroinflammation. 2015. PMID: 25889879 Free PMC article. - Cellular and Molecular Mechanisms of CD8+ T Cell Differentiation, Dysfunction and Exhaustion.
Verdon DJ, Mulazzani M, Jenkins MR. Verdon DJ, et al. Int J Mol Sci. 2020 Oct 5;21(19):7357. doi: 10.3390/ijms21197357. Int J Mol Sci. 2020. PMID: 33027962 Free PMC article. Review. - NFAT signalling is a novel target of oncogenic BRAF in metastatic melanoma.
Flockhart RJ, Armstrong JL, Reynolds NJ, Lovat PE. Flockhart RJ, et al. Br J Cancer. 2009 Oct 20;101(8):1448-55. doi: 10.1038/sj.bjc.6605277. Epub 2009 Sep 1. Br J Cancer. 2009. PMID: 19724275 Free PMC article. - Proteolytically Activated CRAC Effectors through Designed Intramolecular Inhibition.
Jazbec V, Jerala R, Benčina M. Jazbec V, et al. ACS Synth Biol. 2022 Aug 19;11(8):2756-2765. doi: 10.1021/acssynbio.2c00151. Epub 2022 Jul 8. ACS Synth Biol. 2022. PMID: 35802180 Free PMC article. - Calcineurin-Crz1 signaling in lower eukaryotes.
Thewes S. Thewes S. Eukaryot Cell. 2014 Jun;13(6):694-705. doi: 10.1128/EC.00038-14. Epub 2014 Mar 28. Eukaryot Cell. 2014. PMID: 24681686 Free PMC article. Review.
References
- Agarwal S., Avni,O. and Rao,A. (2000) Cell type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity, 12, 1–20. - PubMed
- Anderson G., Moore,N.C., Owen,J.J. and Jenkinson,E.J. (1996) Cellular interactions in thymocyte development. Annu. Rev. Immunol., 14, 73–99. - PubMed
- Aramburu J., Yaffe,M.B., Lopez-Rodriguez,C., Cantley,L.C., Hogan,P.G. and Rao,A. (1999) Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science, 285, 2129–2133. - PubMed
- Brunet A. et al. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell, 96, 857–868. - PubMed
- Brunner T. et al. (1995) Cell-autonomous Fas (CD95)/Fas-ligand interaction mediates activation-induced apoptosis in T-cell hybridomas. Nature, 373, 441–444. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous