Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation - PubMed (original) (raw)
Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation
D Fagegaltier et al. EMBO J. 2000.
Abstract
Decoding of UGA selenocysteine codons in eubacteria is mediated by the specialized elongation factor SelB, which conveys the charged tRNA(Sec) to the A site of the ribosome, through binding to the SECIS mRNA hairpin. In an attempt to isolate the eukaryotic homolog of SelB, a database search in this work identified a mouse expressed sequence tag containing the complete cDNA encoding a novel protein of 583 amino acids, which we called mSelB. Several lines of evidence enabled us to establish that mSelB is the bona fide mammalian elongation factor for selenoprotein translation: it binds GTP, recognizes the Sec-tRNA(Sec) in vitro and in vivo, and is required for efficient selenoprotein translation in vivo. In contrast to the eubacterial SelB, the recombinant mSelB alone is unable to bind specifically the eukaryotic SECIS RNA hairpin. However, complementation with HeLa cell extracts led to the formation of a SECIS-dependent complex containing mSelB and at least another factor. Therefore, the role carried out by a single elongation factor in eubacterial selenoprotein translation is devoted to two or more specialized proteins in eukaryotes.
Figures
Fig. 1. Alignment of SelB sequences from mouse (mSelB), human (hSelB), C.elegans (CeSelB), Drosophila (dSelB), M.jannaschii (MjSelB), E.coli (EcSelB) and of the human hEF1-α sequence. The alignment was made with ClustalW (Thompson et al., 1994) and manually refined with MegAlign (DNASTAR). Identical amino acids are in reverse, similar residues are shaded in gray. The G1–G4 GTP-binding domains are indicated, as well as the Δ1–Δ5 deletions mentioned in the text. The open bar depicts the mSelB G503–I519 block of homology, and the solid bar maps the hSelB FSIK sequence (positions 162–165). The closed circles and the asterisk position residues that are mentioned in the Discussion.
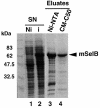
Fig. 2. Purification steps of the recombinant mouse SelB. Supernatants (SN) of E.coli transformed with the His-tagged mSelB expression vector, induced (i in lane 2) or non-induced (Ni in lane 1) by IPTG, were fractionated by affinity chromatography on an Ni-NTA column (lane 3). The eluted fractions were pooled and loaded onto a carboxymethyl-Sephadex C50 column (lane 4). Samples were run on a 10% SDS–polyacrylamide gel and the proteins revealed by Coomassie staining.
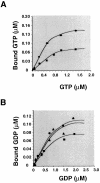
Fig. 3. Determination of the apparent dissociation constants of mSelB for GTP and GDP. Binding assays were performed as described in Materials and methods. (A) The concentration-dependent binding of [3H]GTP to 0.5 (circles) or 0.3 µM (squares) mSelB. (B) Concentration- dependent binding of [3H]GDP to 0.3 (squares), 0.6 (triangles) or 0.9 µM (circles) mSelB.
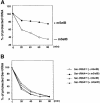
Fig. 4. mSelB specifically protects the selenocysteyl-tRNASec ester bond against mild alkaline hydrolysis. (A) Hydrolysis of the selenocysteyl-tRNASec in the presence (closed circles) or absence (open circles) of mSelB. (B) Hydrolysis of the seryl-tRNASec in the absence (open triangles) or presence (closed triangles) of mSelB. Hydrolysis of the seryl-tRNASer was performed likewise in the absence (open circles) or presence (closed circles) of mSelB.
Fig. 5. The tRNASec is associated with mSelB in vivo. (A) Extracts of COS-7 cells transfected with the HA-tagged mSelB expression vector, or mock-transfected, were blotted with the anti-HA antibody before (lanes 3 and 1, respectively) or after immunoprecipitation (IPP in lanes 4 and 2, respectively). The signal in lane 1 results from cross-reaction of the anti-HA with a cellular protein. Samples were run on a 10% SDS–polyacrylamide gel and revealed by chemiluminescence. (B) Enzymatic determination of the tRNASec sequence with RNase T1 (G), RNase U2 (A), RNase PhyM (A/U) and RNase CL3 (C>U). L is an alkaline ladder; control is a lane that received no enzyme. Shown on the left are sequencing gels (two separate migrations) for the immunoprecipitated tRNASec arising from the experiment in (A). The T7 tRNASec transcript was sequenced in parallel for band assignments. The two-dimensional structure of the mammalian tRNASec (Sturchler et al., 1993) is represented with the modified nucleotides.
Fig. 5. The tRNASec is associated with mSelB in vivo. (A) Extracts of COS-7 cells transfected with the HA-tagged mSelB expression vector, or mock-transfected, were blotted with the anti-HA antibody before (lanes 3 and 1, respectively) or after immunoprecipitation (IPP in lanes 4 and 2, respectively). The signal in lane 1 results from cross-reaction of the anti-HA with a cellular protein. Samples were run on a 10% SDS–polyacrylamide gel and revealed by chemiluminescence. (B) Enzymatic determination of the tRNASec sequence with RNase T1 (G), RNase U2 (A), RNase PhyM (A/U) and RNase CL3 (C>U). L is an alkaline ladder; control is a lane that received no enzyme. Shown on the left are sequencing gels (two separate migrations) for the immunoprecipitated tRNASec arising from the experiment in (A). The T7 tRNASec transcript was sequenced in parallel for band assignments. The two-dimensional structure of the mammalian tRNASec (Sturchler et al., 1993) is represented with the modified nucleotides.
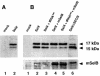
Fig. 6. mSelB is required for efficient selenoprotein translation. (A) Separation of the 17 and 15 kDa SelX polypeptides (lane 2, with 3 µg of SelX expression vector). (B) Fractionation of the SelX polypeptides arising from cells transfected with an excess (10 µg) of SelX expression vector (lane 2), and co-transfected with the tRNASec gene (lane 3). Under the same conditions, co-transfection of the mSelB expression vector is shown, in the absence (lane 4) or presence (lane 5) of the tRNASec gene. Lane 6: expression of SelX in cells transfected with a cDNA lacking the SECIS element. Lanes 1 in (A) and (B) are controls with mock-transfected cells. Proteins were fractionated by 12% SDS–PAGE, blotted with the anti-HA antibody and revealed with the ECL kit. Bottom panel: same extracts as above on a separate (10%) gel where mSelB in lanes 4 and 5 was revealed with the anti-mSelB anti-peptide antibody.
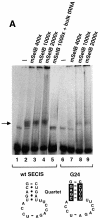
Fig. 7. A protein from HeLa whole-cell extracts forms with mSelB a SECIS-dependent complex. (A) Gel retardation assays obtained with the wild-type GPx SECIS RNA alone (lane 1) or in the presence of increasing amounts of recombinant mSelB protein (400-, 1000- and 2000-fold molar excess mSelB/SECIS RNA, corresponding to 0.4, 1 and 2 µg in lanes 2–4, respectively). Lane 5 contained 700 ng of total yeast tRNA (2-fold molar excess/mSelB). Lanes 6–9 used the G24 GPx SECIS mutant depicted at the bottom. The non-Watson–Crick base pair quartet of the SECIS element is displayed. (B) The wild-type SECIS was incubated alone (lane 1), or in the presence of a 1000-fold molar excess of mSelB alone or complemented with 600 ng of fractionated whole-cell extract (lanes 3 and 4, respectively). The IgG fraction containing the anti-mSelB anti-peptide antiboby or the pre-immune IgGs (PI) were added in lanes 5 and 6, respectively. The G24 SECIS mutant was used in lanes 7–9 under the same conditions as in lanes 1, 3 and 4. Asterisks denote unspecific complexes. WCE, whole-cell extract.
Fig. 7. A protein from HeLa whole-cell extracts forms with mSelB a SECIS-dependent complex. (A) Gel retardation assays obtained with the wild-type GPx SECIS RNA alone (lane 1) or in the presence of increasing amounts of recombinant mSelB protein (400-, 1000- and 2000-fold molar excess mSelB/SECIS RNA, corresponding to 0.4, 1 and 2 µg in lanes 2–4, respectively). Lane 5 contained 700 ng of total yeast tRNA (2-fold molar excess/mSelB). Lanes 6–9 used the G24 GPx SECIS mutant depicted at the bottom. The non-Watson–Crick base pair quartet of the SECIS element is displayed. (B) The wild-type SECIS was incubated alone (lane 1), or in the presence of a 1000-fold molar excess of mSelB alone or complemented with 600 ng of fractionated whole-cell extract (lanes 3 and 4, respectively). The IgG fraction containing the anti-mSelB anti-peptide antiboby or the pre-immune IgGs (PI) were added in lanes 5 and 6, respectively. The G24 SECIS mutant was used in lanes 7–9 under the same conditions as in lanes 1, 3 and 4. Asterisks denote unspecific complexes. WCE, whole-cell extract.
Similar articles
- Protein factors mediating selenoprotein synthesis.
Lescure A, Fagegaltier D, Carbon P, Krol A. Lescure A, et al. Curr Protein Pept Sci. 2002 Feb;3(1):143-51. doi: 10.2174/1389203023380783. Curr Protein Pept Sci. 2002. PMID: 12370018 Review. - Recognition of the mRNA selenocysteine insertion sequence by the specialized translational elongation factor SELB.
Ringquist S, Schneider D, Gibson T, Baron C, Böck A, Gold L. Ringquist S, et al. Genes Dev. 1994 Feb 1;8(3):376-85. doi: 10.1101/gad.8.3.376. Genes Dev. 1994. PMID: 8314089 - Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB.
Kromayer M, Wilting R, Tormay P, Böck A. Kromayer M, et al. J Mol Biol. 1996 Oct 4;262(4):413-20. doi: 10.1006/jmbi.1996.0525. J Mol Biol. 1996. PMID: 8893853 - Distinctive features in the SelB family of elongation factors for selenoprotein synthesis. A glimpse of an evolutionary complexified translation apparatus.
Fagegaltier D, Carbon P, Krol A. Fagegaltier D, et al. Biofactors. 2001;14(1-4):5-10. doi: 10.1002/biof.5520140102. Biofactors. 2001. PMID: 11568434 Review.
Cited by
- The selenocysteine-specific elongation factor contains a novel and multi-functional domain.
Gonzalez-Flores JN, Gupta N, DeMong LW, Copeland PR. Gonzalez-Flores JN, et al. J Biol Chem. 2012 Nov 9;287(46):38936-45. doi: 10.1074/jbc.M112.415463. Epub 2012 Sep 19. J Biol Chem. 2012. PMID: 22992746 Free PMC article. - Characterization of the UGA-recoding and SECIS-binding activities of SECIS-binding protein 2.
Bubenik JL, Miniard AC, Driscoll DM. Bubenik JL, et al. RNA Biol. 2014;11(11):1402-13. doi: 10.1080/15476286.2014.996472. RNA Biol. 2014. PMID: 25692238 Free PMC article. - Threading the needle: getting selenocysteine into proteins.
Donovan J, Copeland PR. Donovan J, et al. Antioxid Redox Signal. 2010 Apr 1;12(7):881-92. doi: 10.1089/ars.2009.2878. Antioxid Redox Signal. 2010. PMID: 19747061 Free PMC article. Review. - Crystal structure analysis reveals functional flexibility in the selenocysteine-specific tRNA from mouse.
Ganichkin OM, Anedchenko EA, Wahl MC. Ganichkin OM, et al. PLoS One. 2011;6(5):e20032. doi: 10.1371/journal.pone.0020032. Epub 2011 May 24. PLoS One. 2011. PMID: 21629646 Free PMC article. - Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities.
Lubos E, Loscalzo J, Handy DE. Lubos E, et al. Antioxid Redox Signal. 2011 Oct 1;15(7):1957-97. doi: 10.1089/ars.2010.3586. Epub 2011 Apr 10. Antioxid Redox Signal. 2011. PMID: 21087145 Free PMC article. Review.
References
- Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) Dynamics of the genetic code. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 637–673.
- Berry M.J., Banu,L., Chen,Y.Y., Mandel,S.J., Kieffer,J.D., Harney,J.W. and Larsen,P.R. (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ UTR. Nature, 353, 273–276. - PubMed
- Buettner C., Harney,J.W. and Berry,M.J. (1999) The Caenorhabditis elegans homologue of thioredoxin reductase contains a SECIS element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J. Biol. Chem., 274, 21598–21602. - PubMed
- Burk R.F. and Hill,K.E. (1999) Orphan selenoproteins. BioEssays, 21, 231–237. - PubMed
- Commans S. and Böck,A. (1999) Selenocysteine inserting tRNAs: an overview. FEMS Microbiol. Rev., 23, 335–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases