Replication-dependent early meiotic requirement for Spo11 and Rad50 - PubMed (original) (raw)
Replication-dependent early meiotic requirement for Spo11 and Rad50
S T Merino et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Spo11 and the Rad50-Mre11 complex have been indirectly implicated in processes associated with DNA replication. These proteins also have been shown to have early meiotic roles essential for the formation of a programmed DNA double-strand break known in Saccharomyces cerevisiae to initiate meiotic recombination. In both S. cerevisiae and the basidiomycete Coprinus cinereus, spo11 and rad50 mutants are defective in chromosome synapsis during meiosis. Here we demonstrate that a partial restoration of synapsis occurs in C. cinereus spo11 and rad50 mutants if premeiotic DNA replication is prevented. Double mutants were constructed with spo11-1 or rad50-4 and another mutant, spo22-1, which does not undergo premeiotic DNA replication. In both cases, we observed an increase in the percentage of nuclei containing synaptonemal complex (SC) structures, with concomitant decreases in the percentage of nuclei containing axial elements (AE) only or no structures. Both types of double mutants demonstrated significant increases in the average numbers of AE and SC, although SC-containing nuclei did not on average contain more AE than did nuclei showing no synapsis. Our results show that Spo11-induced recombination is not absolutely required for synapsis in C. cinereus, and that the early meiotic role of both Spo11 and Rad50 in SC formation partially depends on premeiotic S phase. This dependency likely reflects either a requirement for these proteins imposed by the premeiotic replication process itself or a requirement for these proteins in synapsis when a sister chromatid (the outcome of DNA replication) is present.
Figures
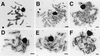
Figure 1
SC formation in wild-type and mutant strains. (A) TSM1, wild type; (B) TSM3, spo11–1; (C) TSM4, rad50–4; (D) TSM2, spo22–1; (E) TSM5, spo11–1;spo22–1; and (F) TSM8, rad50–4;spo22–1. [Scale bars = 2 μm.]
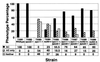
Figure 2
Comparison of wild-type, single-, and double-mutant strains. Chromosome spreads were examined and categorized according to the presence or absence of AE and SC. For each strain, at least two mushrooms were sampled. Total numbers of spreads were: TSM1, 22; TSM2, 19; TSM3, 60; TSM4, 30; TSM5, 27; TSM6, 45; TSM7, 38; TSM8, 40; and TSM9, 27.
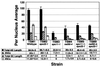
Figure 3
Comparison of SC and AE length vs. the number of SC and AE segments. Total SC and AE lengths were measured and the sum was divided by the total number of nuclei to determine the average length per nucleus for each strain listed in the table. For each chromosome spread, AE segments were counted as one segment for each continuous piece regardless of length. A single SC segment represents continuous association of two AE. Data are presented as ± indicates the SEM. SC and AE lengths are in micrometers (μm).
Similar articles
- Multiple roles of Spo11 in meiotic chromosome behavior.
Celerin M, Merino ST, Stone JE, Menzie AM, Zolan ME. Celerin M, et al. EMBO J. 2000 Jun 1;19(11):2739-50. doi: 10.1093/emboj/19.11.2739. EMBO J. 2000. PMID: 10835371 Free PMC article. - Coprinus cinereus rad50 mutants reveal an essential structural role for Rad50 in axial element and synaptonemal complex formation, homolog pairing and meiotic recombination.
Acharya SN, Many AM, Schroeder AP, Kennedy FM, Savytskyy OP, Grubb JT, Vincent JA, Friedle EA, Celerin M, Maillet DS, Palmerini HJ, Greischar MA, Moncalian G, Williams RS, Tainer JA, Zolan ME. Acharya SN, et al. Genetics. 2008 Dec;180(4):1889-907. doi: 10.1534/genetics.108.092775. Epub 2008 Oct 20. Genetics. 2008. PMID: 18940790 Free PMC article. - Meiotic localization of Mre11 and Rad50 in wild type, spo11-1, and MRN complex mutants of Coprinus cinereus.
Many AM, Melki CS, Savytskyy OP, Maillet DS, Acharya SN, Zolan ME. Many AM, et al. Chromosoma. 2009 Aug;118(4):471-86. doi: 10.1007/s00412-009-0209-5. Epub 2009 Apr 26. Chromosoma. 2009. PMID: 19396455 - [SPO11: an activity that promotes DNA breaks required for meiosis].
Baudat F, de Massy B. Baudat F, et al. Med Sci (Paris). 2004 Feb;20(2):213-8. doi: 10.1051/medsci/2004202213. Med Sci (Paris). 2004. PMID: 14997442 Review. French. - [Latest frontiers of meiosis research in Coprinus cinereus].
Namekawa SH, Hamada FN, Sakaguchi K. Namekawa SH, et al. Seikagaku. 2004 Nov;76(11):1450-4. Seikagaku. 2004. PMID: 15626033 Review. Japanese. No abstract available.
Cited by
- Comparative transcriptomics of the model mushroom Coprinopsis cinerea reveals tissue-specific armories and a conserved circuitry for sexual development.
Plaza DF, Lin CW, van der Velden NS, Aebi M, Künzler M. Plaza DF, et al. BMC Genomics. 2014 Jun 19;15(1):492. doi: 10.1186/1471-2164-15-492. BMC Genomics. 2014. PMID: 24942908 Free PMC article. - Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2.
MacQueen AJ, Villeneuve AM. MacQueen AJ, et al. Genes Dev. 2001 Jul 1;15(13):1674-87. doi: 10.1101/gad.902601. Genes Dev. 2001. PMID: 11445542 Free PMC article. - Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice.
Kumar R, Bourbon HM, de Massy B. Kumar R, et al. Genes Dev. 2010 Jun 15;24(12):1266-80. doi: 10.1101/gad.571710. Genes Dev. 2010. PMID: 20551173 Free PMC article. - Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans.
Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. Jantsch V, et al. Mol Cell Biol. 2004 Sep;24(18):7998-8006. doi: 10.1128/MCB.24.18.7998-8006.2004. Mol Cell Biol. 2004. PMID: 15340062 Free PMC article. - Differentiated function and localisation of SPO11-1 and PRD3 on the chromosome axis during meiotic DSB formation in Arabidopsis thaliana.
Lambing C, Kuo P, Kim J, Osman K, Whitbread AL, Yang J, Choi K, Franklin FCH, Henderson IR. Lambing C, et al. PLoS Genet. 2022 Jul 20;18(7):e1010298. doi: 10.1371/journal.pgen.1010298. eCollection 2022 Jul. PLoS Genet. 2022. PMID: 35857772 Free PMC article.
References
- Moore D P, Orr-Weaver T L. Curr Top Dev Biol. 1998;37:263–299. - PubMed
- Malone R E. Mol Gen Genet. 1983;189:405–412. - PubMed
- Petes T D, Malone R E, Symington L S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces, Genome Dynamics, Protein Synthesis, and Energetics. Broach J R, Pringle J R, Jones E W, editors. New York: Cold Spring Harbor Lab. Press; 1991. pp. 407–521.
- Keeney S, Giroux C N, Kleckner N. Cell. 1997;88:375–384. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous