Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules - PubMed (original) (raw)
Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules
R K Miller et al. Mol Biol Cell. 2000 Sep.
Free PMC article
Abstract
In Saccharomyces cerevisiae, positioning of the mitotic spindle depends on the interaction of cytoplasmic microtubules with the cell cortex. In this process, cortical Kar9p in the bud acts as a link between the actin and microtubule cytoskeletons. To identify Kar9p-interacting proteins, a two-hybrid screen was conducted with the use of full-length Kar9p as bait, and three genes were identified: BIM1, STU2, and KAR9 itself. STU2 encodes a component of the spindle pole body. Bim1p is the yeast homologue of the human microtubule-binding protein EB1, which is a binding partner to the adenomatous polyposis coli protein involved in colon cancer. Eighty-nine amino acids within the third quarter of Bim1p was sufficient to confer interaction with Kar9p. The two-hybrid interactions were confirmed with the use of coimmunoprecipitation experiments. Genetic analysis placed Bim1p in the Kar9p pathway for nuclear migration. Bim1p was not required for Kar9p's cortical or spindle pole body localization. However, deletion of BIM1 eliminated Kar9p localization along cytoplasmic microtubules. Furthermore, in the bim1 mutants, the cytoplasmic microtubules no longer intersected the cortical dot of Green Fluorescent Protein-Kar9p. These experiments demonstrate that the interaction of cytoplasmic microtubules with the Kar9p cortical attachment site requires the microtubule-binding protein Bim1p.
Figures
Figure 1
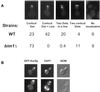
Kar9p interacts with Bim1p, Stu2p, and Kar9p by two-hybrid analysis. (A) The wild-type two-hybrid strain (PJ69-4A) containing pGDB-KAR9 (pMR4150) and the indicated pGAD-BIM1 library-positive plasmids were serially diluted 10-fold and spotted onto plates lacking uracil and leucine (−ura leu) or adenine (−ade). Plates were photographed after 2 d at 30°C. (B) A two-hybrid reporter strain deleted for_BIM1_ (MY7309) containing either GDB-KAR9 (pMR4150) or empty GDB vector (pMR3763) was transformed with the indicated STU2 plasmids and assayed for activation of the HIS3 reporter gene. The microtubule-binding domain of Stu2p (MT) has been mapped to amino acids 558–658 (Wang and Huffaker, 1997). Three independent STU2 transformants are shown. Plates were photographed after 3 d at 30°C. (C) The wild-type two-hybrid strain (PJ69-4A) containing pGDB-KAR9 (pMR4150) and the indicated_KAR9_ library plasmids were serially diluted 10-fold and spotted onto plates lacking uracil and leucine (−ura leu) or adenine (−ade). Plates were photographed after 2 d at 30°C. (D and E) The Genetics Computer Group (Madison, WI) programs Plotsimilarity and Pretty were used to compare the EB1 homologues from human (accession number I52726), mouse (accession number AAA96320), catfish (accession number AAD31035), D. melanogaster (accession number AAD27859), urochordate (accession number CAA67697), C. elegans (accession number CAA20332), S. pombe (accession number Q10113), and S. cerevisiae (accession number NP 010932). (E) The BLOSUM62 amino acid substitution matrix was used for calculations. Additional parameters were set as GapLengthWeight, 2; GapWeight, 8. A consensus was noted if six or more residues were similar. The location of the Kar9p-binding domain is indicated by closed circles.
Figure 1
Kar9p interacts with Bim1p, Stu2p, and Kar9p by two-hybrid analysis. (A) The wild-type two-hybrid strain (PJ69-4A) containing pGDB-KAR9 (pMR4150) and the indicated pGAD-BIM1 library-positive plasmids were serially diluted 10-fold and spotted onto plates lacking uracil and leucine (−ura leu) or adenine (−ade). Plates were photographed after 2 d at 30°C. (B) A two-hybrid reporter strain deleted for_BIM1_ (MY7309) containing either GDB-KAR9 (pMR4150) or empty GDB vector (pMR3763) was transformed with the indicated STU2 plasmids and assayed for activation of the HIS3 reporter gene. The microtubule-binding domain of Stu2p (MT) has been mapped to amino acids 558–658 (Wang and Huffaker, 1997). Three independent STU2 transformants are shown. Plates were photographed after 3 d at 30°C. (C) The wild-type two-hybrid strain (PJ69-4A) containing pGDB-KAR9 (pMR4150) and the indicated_KAR9_ library plasmids were serially diluted 10-fold and spotted onto plates lacking uracil and leucine (−ura leu) or adenine (−ade). Plates were photographed after 2 d at 30°C. (D and E) The Genetics Computer Group (Madison, WI) programs Plotsimilarity and Pretty were used to compare the EB1 homologues from human (accession number I52726), mouse (accession number AAA96320), catfish (accession number AAD31035), D. melanogaster (accession number AAD27859), urochordate (accession number CAA67697), C. elegans (accession number CAA20332), S. pombe (accession number Q10113), and S. cerevisiae (accession number NP 010932). (E) The BLOSUM62 amino acid substitution matrix was used for calculations. Additional parameters were set as GapLengthWeight, 2; GapWeight, 8. A consensus was noted if six or more residues were similar. The location of the Kar9p-binding domain is indicated by closed circles.
Figure 2
Kar9p coimmunoprecipitates with Bim1p, Stu2p, and Kar9p. (A) A kar9_Δ stain (MS4263) containing HA epitope–tagged KAR9 (pMR4723), nontagged_KAR9 (pMR4722), or galactose-inducible_BIM1_::V5 (pMR4620) plasmids were grown to mid exponential phase, and extracts were prepared for immunoprecipitation as described in MATERIALS AND METHODS. V5 antibody conjugated to Sepharose beads was used to immunoprecipitate Bim1p::V5. It was also used to probe Western blots. The HA antibody HA.11(16B12) conjugated to agarose beads was used to immunoprecipitate Kar9p::HA and probe the resulting Western blot. The HA antibody 12CA5 was used as a probe in the V5 immunoprecipitation experiment. Identical results were obtained when the two HA antibodies were used in any combination for either the immunoprecipitation step or subsequent Western blots. (B) Stu2p::HA was immunoprecipitated from extracts prepared from wild-type (MS1556) or_kar9_Δ (MS4306) strains containing HA epitope–tagged_STU2_ (pWP70) or empty vector (pRS415; Sikorski and Hieter, 1989), as described in MATERIALS AND METHODS. Proteins from the precipitates were analyzed by Western blotting with the use of rabbit anti-Kar9p and the HA antibody 12CA5. (C) GFP–Kar9p was immunoprecipitated with the use of anti-GFP (a kind gift from Pamela Silver) from wild-type strains transformed with Kar9p:HA (pMR4723), Kar9p with no tag (pMR4722), and PGAL1-GFP-Kar9p (pMR3464). Proteins from the precipitates were analyzed by Western blotting with the use of rabbit anti-Kar9p and the HA antibody 12CA5.
Figure 3
Bim1p is required for localization of GFP–Kar9p along microtubules but not for Kar9p localization at the SPB or cortex. (A) Wild-type (MS1556) and _bim1_Δ (MS7310) strains containing GFP–Kar9p (pMR3465) were grown to early exponential phase and induced for GFP-Kar9p expression. The percentage of cells exhibiting GFP-Kar9p expression in medium to large-budded cells with a single nucleus is shown under each representative cell (n = 720 for wild-type cells and 660 for _bim1_Δ cells). The average of three independent experiments is shown. Five percent of wild-type and 6% of _bim1_Δ cells exhibited GFP–Kar9p localization patterns that are not depicted. (B) A_bim1_Δ strain (MS7310) containing GFP–Kar9p (pMR3465) was grown and prepared as described in A.
Figure 4
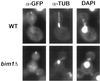
Cytoplasmic microtubules do not intersect the Kar9p dot in _bim1_Δ cells. Wild-type and_bim1_Δ cells containing GFP–Kar9p were fixed for double-label indirect immunofluorescence with the use of anti-GFP, anti-tubulin, and DAPI, as described in MATERIALS AND METHODS (n = 96 for wild-type cells and 105 for _bim1_Δ cells).
Similar articles
- The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization.
Miller RK, Matheos D, Rose MD. Miller RK, et al. J Cell Biol. 1999 Mar 8;144(5):963-75. doi: 10.1083/jcb.144.5.963. J Cell Biol. 1999. PMID: 10085294 Free PMC article. - Positioning of the mitotic spindle by a cortical-microtubule capture mechanism.
Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Lee L, et al. Science. 2000 Mar 24;287(5461):2260-2. doi: 10.1126/science.287.5461.2260. Science. 2000. PMID: 10731147 - BIM1 encodes a microtubule-binding protein in yeast.
Schwartz K, Richards K, Botstein D. Schwartz K, et al. Mol Biol Cell. 1997 Dec;8(12):2677-91. doi: 10.1091/mbc.8.12.2677. Mol Biol Cell. 1997. PMID: 9398684 Free PMC article. - Search, capture and signal: games microtubules and centrosomes play.
Schuyler SC, Pellman D. Schuyler SC, et al. J Cell Sci. 2001 Jan;114(Pt 2):247-55. doi: 10.1242/jcs.114.2.247. J Cell Sci. 2001. PMID: 11148127 Review. - Microtubule cytoskeleton: navigating the intracellular landscape.
Bloom K. Bloom K. Curr Biol. 2003 May 27;13(11):R430-2. doi: 10.1016/s0960-9822(03)00362-2. Curr Biol. 2003. PMID: 12781150 Review.
Cited by
- The MEN mediates the effects of the spindle assembly checkpoint on Kar9-dependent spindle pole body inheritance in budding yeast.
Hotz M, Lengefeld J, Barral Y. Hotz M, et al. Cell Cycle. 2012 Aug 15;11(16):3109-16. doi: 10.4161/cc.21504. Epub 2012 Aug 8. Cell Cycle. 2012. PMID: 22871738 Free PMC article. - Overlap of cargo binding sites on myosin V coordinates the inheritance of diverse cargoes.
Eves PT, Jin Y, Brunner M, Weisman LS. Eves PT, et al. J Cell Biol. 2012 Jul 9;198(1):69-85. doi: 10.1083/jcb.201201024. Epub 2012 Jul 2. J Cell Biol. 2012. PMID: 22753895 Free PMC article. - The TOG protein Stu2 is regulated by acetylation.
Greenlee MA, Witt B, Sabo JA, Morris SC, Miller RK. Greenlee MA, et al. PLoS Genet. 2022 Sep 9;18(9):e1010358. doi: 10.1371/journal.pgen.1010358. eCollection 2022 Sep. PLoS Genet. 2022. PMID: 36084134 Free PMC article. - Multivalency ensures persistence of a +TIP body at specialized microtubule ends.
Meier SM, Farcas AM, Kumar A, Ijavi M, Bill RT, Stelling J, Dufresne ER, Steinmetz MO, Barral Y. Meier SM, et al. Nat Cell Biol. 2023 Jan;25(1):56-67. doi: 10.1038/s41556-022-01035-2. Epub 2022 Dec 19. Nat Cell Biol. 2023. PMID: 36536177 Free PMC article. - Monitoring spindle orientation: Spindle position checkpoint in charge.
Caydasi AK, Ibrahim B, Pereira G. Caydasi AK, et al. Cell Div. 2010 Dec 11;5:28. doi: 10.1186/1747-1028-5-28. Cell Div. 2010. PMID: 21143992 Free PMC article.
References
- Barker N, Morin PJ, Clevers H. The yin-yang of TCF/beta-catenin signaling. Adv Cancer Res. 2000;77:1–24. - PubMed
- Barth AI, Nathke IS, Nelson WJ. Cadherins, catenins and APC protein: interplay between cytoskeletal complexes and signaling pathways. Curr Opin Cell Biol. 1997;9:683–690. - PubMed
- Byers B. Cytology of the yeast life cycle. In: Strathern J, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 59–96.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases