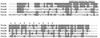Analysis of the essential cell division gene ftsL of Bacillus subtilis by mutagenesis and heterologous complementation - PubMed (original) (raw)
Analysis of the essential cell division gene ftsL of Bacillus subtilis by mutagenesis and heterologous complementation
J Sievers et al. J Bacteriol. 2000 Oct.
Abstract
The ftsL gene is required for the initiation of cell division in a broad range of bacteria. Bacillus subtilis ftsL encodes a 13-kDa protein with a membrane-spanning domain near its N terminus. The external C-terminal domain has features of an alpha-helical leucine zipper, which is likely to be involved in the heterodimerization with another division protein, DivIC. To determine what residues are important for FtsL function, we used both random and site-directed mutagenesis. Unexpectedly, all chemically induced mutations fell into two clear classes, those either weakening the ribosome-binding site or producing a stop codon. It appears that the random mutagenesis was efficient, so many missense mutations must have been generated but with no phenotypic effect. Substitutions affecting hydrophobic residues in the putative coiled-coil domain, introduced by site-directed mutagenesis, also gave no observable phenotype except for insertion of a helix-breaking proline residue, which destroyed FtsL function. ftsL homologues cloned from three diverse Bacillus species, Bacillus licheniformis, Bacillus badius, and Bacillus circulans, could complement an ftsL null mutation in B. subtilis, even though up to 66% of the amino acid residues of the predicted proteins were different from B. subtilis FtsL. However, the ftsL gene from Staphylococcus aureus (whose product has 73% of its amino acids different from those of the B. subtilis ftsL product) was not functional. We conclude that FtsL is a highly malleable protein that can accommodate a large number of sequence changes without loss of function.
Figures
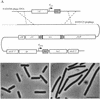
FIG. 1
Strain 2007 constructed for mutagenesis of ftsL. (A) Schematic representation of the genetic organization of strain 2007. The chromosomal pbpB operon contains four genes, yllB, yllC, ftsL, and pbpB. The chromosomal copy of ftsL is disrupted by a neo cassette, whose promoter also provides transcription of the downstream pbpB gene. An IPTG-inducible wild-type copy of ftsL is placed in the amyE locus. The φ105J125 prophage, integrated elsewhere in the chromosome, allows transformation with φ105J506 phage DNA and the insertion by a double-crossover event (dashed lines) of a second copy of ftsL (ftsL*) under the control of the xylose-inducible Pxyl promoter. (B and C) Phase-contrast images of 2007 grown in the presence of the inducer IPTG (B) and after ca. three generations in its absence (C). Bar, 5 μm.
FIG. 2
Comparison of the FtsL proteins of B. subtilis (FtsLBs), B. licheniformis (FtsLBl), B. badius (FtsLBb), B. circulans (FtsLBc), and S. aureus (FtsLSa). The alignment was performed using the CLUSTAL W method, with residues identical to the B. subtilis sequence shown in gray boxes. The membrane-spanning domain and the a and d positions of the heptad repeat of the putative coiled-coil domain are shown above the alignment.
FIG. 3
Complementation of a B. subtilis ftsL null mutation by ftsL alleles of S. aureus, B. licheniformis, B. circulans, and B. badius. Shown are phase-contrast images of strains 2044, 2045, 2046, and 2047 grown at 37°C in PAB supplemented with either IPTG (left panel) or xylose (central panel) or not supplemented with any inducer (right panel). IPTG induces the expression of the wild-type gene, whereas the foreign ftsL alleles are transcribed in the presence of xylose. As expected, all four strains were filamented in unsupplemented medium (i.e., when FtsL is depleted). Bar, 5 μm.
Similar articles
- Characterization of the essential cell division gene ftsL(yIID) of Bacillus subtilis and its role in the assembly of the division apparatus.
Daniel RA, Harry EJ, Katis VL, Wake RG, Errington J. Daniel RA, et al. Mol Microbiol. 1998 Jul;29(2):593-604. doi: 10.1046/j.1365-2958.1998.00954.x. Mol Microbiol. 1998. PMID: 9720875 - Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis.
Bramkamp M, Weston L, Daniel RA, Errington J. Bramkamp M, et al. Mol Microbiol. 2006 Oct;62(2):580-91. doi: 10.1111/j.1365-2958.2006.05402.x. Mol Microbiol. 2006. PMID: 17020588 - Cell division in Escherichia coli: role of FtsL domains in septal localization, function, and oligomerization.
Ghigo JM, Beckwith J. Ghigo JM, et al. J Bacteriol. 2000 Jan;182(1):116-29. doi: 10.1128/JB.182.1.116-129.2000. J Bacteriol. 2000. PMID: 10613870 Free PMC article. - FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli.
Guzman LM, Barondess JJ, Beckwith J. Guzman LM, et al. J Bacteriol. 1992 Dec;174(23):7716-28. J Bacteriol. 1992. PMID: 1332942 Free PMC article. - The membrane: transertion as an organizing principle in membrane heterogeneity.
Matsumoto K, Hara H, Fishov I, Mileykovskaya E, Norris V. Matsumoto K, et al. Front Microbiol. 2015 Jun 12;6:572. doi: 10.3389/fmicb.2015.00572. eCollection 2015. Front Microbiol. 2015. PMID: 26124753 Free PMC article. Review.
Cited by
- Proline substitutions in a Mip-like peptidyl-prolyl cis-trans isomerase severely affect its structure, stability, shape and activity.
Polley S, Chakravarty D, Chakrabarti G, Chattopadhyaya R, Sau S. Polley S, et al. Biochim Open. 2015 Jul 23;1:28-39. doi: 10.1016/j.biopen.2015.07.001. eCollection 2015. Biochim Open. 2015. PMID: 29632827 Free PMC article. - Cell Cycle Machinery in Bacillus subtilis.
Errington J, Wu LJ. Errington J, et al. Subcell Biochem. 2017;84:67-101. doi: 10.1007/978-3-319-53047-5_3. Subcell Biochem. 2017. PMID: 28500523 Free PMC article. Review. - DNA repair and genome maintenance in Bacillus subtilis.
Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. Lenhart JS, et al. Microbiol Mol Biol Rev. 2012 Sep;76(3):530-64. doi: 10.1128/MMBR.05020-11. Microbiol Mol Biol Rev. 2012. PMID: 22933559 Free PMC article. Review. - Role of leucine zipper motifs in association of the Escherichia coli cell division proteins FtsL and FtsB.
Robichon C, Karimova G, Beckwith J, Ladant D. Robichon C, et al. J Bacteriol. 2011 Sep;193(18):4988-92. doi: 10.1128/JB.00324-11. Epub 2011 Jul 22. J Bacteriol. 2011. PMID: 21784946 Free PMC article. - Assembly of the Caulobacter cell division machine.
Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. Goley ED, et al. Mol Microbiol. 2011 Jun;80(6):1680-98. doi: 10.1111/j.1365-2958.2011.07677.x. Epub 2011 May 17. Mol Microbiol. 2011. PMID: 21542856 Free PMC article.
References
- Beall B, Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991;5:447–455. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources