Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms - PubMed (original) (raw)
Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms
T Kahn et al. Mol Cell Biol. 2000 Oct.
Abstract
Drosophila telomeres contain arrays of the retrotransposonlike elements HeT-A and TART. Their transposition to broken chromosomal termini has been implicated in chromosome healing and telomere elongation. The HeT-A element is attached by its 3' end, which contains the promoter. To monitor the behavior of HeT-A elements, we used the yellow gene with terminal deficiencies consisting of breaks in the yellow promoter region that result in the y-null phenotype. Attachment of the HeT-A element provides the promoterless yellow gene with a promoter that activates yellow expression in bristles. The frequency of HeT-A transpositions to the yellow terminal deficiency depends on the genotype of the line and varies from 2 x 10(-3) to less than 2 x 10(-5). Loss of the attached HeT-A due to incomplete replication at the telomere leads to inactivation of yellow expression, which is restored by attachment of a new HeT-A element upstream of yellow. New HeT-A additions occur at a frequency of about 1.2 x 10(-3). Short DNA attachments are generated by gene conversion using the homologous telomeric sequences as templates. Longer DNA attachments are generated either by conventional transposition of an HeT-A element to the chromosomal terminus or by recombination between the 3' terminus of telomeric HeT-A elements and the receding end of HeT-A attached to the yellow gene.
Figures
FIG. 1
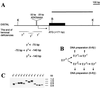
Minimal promoter region responsible for yellow activation at the tip of the terminal deficiencies. (A) A schematic presentation of terminal yellow deficiencies associated with different y phenotypes. The localization of regulatory regions, promoter, and start of translation are indicated according to the transcription start site of the yellow gene. The coding yellow region is shown as a black box. The sequence and localization of the yellow promoter is presented. The start of yellow transcription is shown by an arrow. The translation start codon (ATG) is indicated. The thin horizontal lines show the regions of yellow sequence in which the termini of the yTD line that correspond to the same class of y phenotype have been mapped. A _Bam_HI-_Kpn_I genomic fragment used as a probe for Southern blot analysis is indicated by a thick line in the upper part of the figure. Restriction enzyme abbreviations: B, _Bam_HI; K, _Kpn_I. (B) Scheme used to study the correlation between the DNA structure and y phenotype. In subsequent generations two or three yTD/yac sisters displaying either the y2-like or the yv or y1-like phenotype were crossed individually with yac males. Other females in groups of six to eight were combined according to their phenotypes and used for DNA preparation. (C) Southern blot analysis of DNAs prepared from six to eight yTD/yac sisters displaying either y2-like or yv or y1-like phenotype. DNAs were digested with _Kpn_I. The filter was hybridized with the _Bam_HI-_Kpn_I probe.
FIG. 2
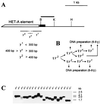
HeT-A transpositions to the broken chromosome terminus in the yellow gene. (A) Partial restriction map of the wild-type yellow gene region, indicating the position of the _Bam_HI-_Kpn_I probe that was used for Southern blot analysis. The coding yellow region is shown as a black box. Restriction enzyme abbreviations: B, _Bam_HI; K, _Kpn_I; N, _Nru_I. The primers in the HeT-A element and the yellow gene used for DNA amplification are shown by arrows. (B) HeT-A additions as indicated by Southern blot hybridization of genomic DNA restricted with _Nru_I and probed with the _Bam_HI-_Kpn_I fragment. The _Nru_I site is at position +1835 relative to the transcription start site of the yellow gene. (C) Attachment points of HeT-A elements in the yellow gene. The start of yellow transcription is shown by a bent arrow. The translation start codon ATG is underlined. The points of HeT-A attachment are shown by small arrows. The attachment of HeT-A in the yhTD3 line is indicated by a large arrow. One or two A bases at the junctions may originate either from the receding yellow sequences or from the oligo(A) tail of the 3′ end of the attached HeT-A element.
FIG. 3
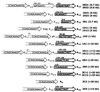
Minimal region of the HeT-A element which is sufficient for compensation of the yellow promoter deletion at the tip of the X chromosome. (A) Scheme of terminal deficiencies associated with different y phenotypes. The yellow coding region is shown by a black box. The thin horizontal lines in the lower part of the scheme show the regions of the HeT-A element (numbered from the 3′ HeT-A end) in which termini that correspond to the y phenotype have been mapped. The figures show the distance from the first nucleotide of the 3′ end of the HeT-A element. Restriction enzyme abbreviation: H, _Sph_I. Other designations are as in Fig. 1. (B) The scheme used to study the relationship between the length of the terminal HeT-A element and the y phenotype. (C) Southern blot analysis of yhTD3/yac sisters displaying y2-like, yv, or y1-like phenotypes. DNAs were digested with _Sph_I. The filter was hybridized with the _Bam_HI-_Kpn_I fragment indicated by the thick line in panel A. The _Sph_I site is 1435 bp from the HeT-A attachment in yhTD3.
FIG. 4
Molecular additions to the tip of the X chromosome. (A) Partial restriction map of the wild-type yellow gene region, indicating the position of sites for restriction enzymes used for Southern blot analysis. The HeT-A addition is shown by a shaded box. The coding yellow regions are shown by black boxes. Restriction enzyme abbreviations: B, _Bam_HI; K, _Kpn_I; E, _Eco_RV; H, _Sph_I; S, _Spe_I; N, _Nru_I; R, _Eco_RI. Other designations are as in Fig. 1 and 2. (B through D) Southern blot analysis of DNAs prepared from _y2_-like lines. DNAs were digested with _Nru_I (B), _Eco_RI (C), and _Kpn_I (D). The filters were hybridized with the _Bam_HI-_Kpn_I fragment.
FIG. 5
Restriction map of the DNA additions to the terminal HeT-A element as inferred from genomic Southern blot analysis. The maps of the DNA additions obtained as clusters of identical events are linked by vertical lines. The maps start from the _Bam_HI site located in the yellow gene at 39 bp from the HeT-A attachment site in the yhTD3 line. (A) Restriction maps of the DNA attachments with a determined size; (B) restriction maps of the DNA attachments with undetermined size. The lines described in Fig. 6 and 7 are underlined. Abbreviations of restriction enzymes are as in Fig. 4.
FIG. 6
Aligned sequences of the original HeT-A element and four _y2_-like derivatives. All sequences end with the last nucleotide at the 3′ end of the original HeT-A element. The sequences are shown in the 5′-to-3′ orientation. Only the last 350 nucleotides of aligned sequences are shown. The small letters show the substitutions in the sequence. Asterisks indicate missing nucleotides. Arrows represent the 3′ ends of new HeT-A elements. The sequences that may refer to the original HeT-A element in the yhTD3 line are shown by bold letters.
FIG. 7
Diagram of HeT-A additions to the receding HeT-A element. The numbers in parentheses show the approximate sizes of the attachments. The arrows indicate the directions of the HeT-A elements. The bold arrows correspond to the original HeT-A element. The numbers above the arrows indicate distances of either the 5′ terminus or the 5′ and 3′ termini of the HeT-A element from the 3′ terminus of a standard element. For this, the terminal HeT-A element present in the original yhTD3 line was used (Fig. 6). A base at the junctions may originate either from the terminal HeT-A element or from the 3′ oligo(A) tail of the new HeT-A element. The base pairs at the junction between new and old HeT-A elements are shown. The lowercase letters indicate substitutions in the conserved sequence at the 3′ end of the HeT-A element.
Similar articles
- Enhancer of terminal gene conversion, a new mutation in Drosophila melanogaster that induces telomere elongation by gene conversion.
Melnikova L, Georgiev P. Melnikova L, et al. Genetics. 2002 Nov;162(3):1301-12. doi: 10.1093/genetics/162.3.1301. Genetics. 2002. PMID: 12454074 Free PMC article. - [Model genetic system for analysis of attachment of HeT-A elements to terminal deletions in Drosophila melanogaster].
Kan TG, Selezneva TB, Georgiev PG. Kan TG, et al. Genetika. 2000 Nov;36(11):1515-9. Genetika. 2000. PMID: 11094769 Russian. - Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster.
Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Savitsky M, et al. Mol Cell Biol. 2002 May;22(9):3204-18. doi: 10.1128/MCB.22.9.3204-3218.2002. Mol Cell Biol. 2002. PMID: 11940677 Free PMC article. - Drosophila telomere elongation.
Biessmann H, Walter MF, Mason JM. Biessmann H, et al. Ciba Found Symp. 1997;211:53-67; discussion 67-70. doi: 10.1002/9780470515433.ch5. Ciba Found Symp. 1997. PMID: 9524751 Review. - Drosophila: Retrotransposons Making up Telomeres.
Casacuberta E. Casacuberta E. Viruses. 2017 Jul 19;9(7):192. doi: 10.3390/v9070192. Viruses. 2017. PMID: 28753967 Free PMC article. Review.
Cited by
- The Ku protein complex is involved in length regulation of Drosophila telomeres.
Melnikova L, Biessmann H, Georgiev P. Melnikova L, et al. Genetics. 2005 May;170(1):221-35. doi: 10.1534/genetics.104.034538. Epub 2005 Mar 21. Genetics. 2005. PMID: 15781709 Free PMC article. - Transvection at the end of the truncated chromosome in Drosophila melanogaster.
Savitsky M, Kahn T, Pomerantseva E, Georgiev P. Savitsky M, et al. Genetics. 2003 Apr;163(4):1375-87. doi: 10.1093/genetics/163.4.1375. Genetics. 2003. PMID: 12702682 Free PMC article. - The protein encoded by the gene proliferation disrupter (prod) is associated with the telomeric retrotransposon array in Drosophila melanogaster.
Török T, Benitez C, Takács S, Biessmann H. Török T, et al. Chromosoma. 2007 Apr;116(2):185-95. doi: 10.1007/s00412-006-0090-4. Epub 2006 Dec 21. Chromosoma. 2007. PMID: 17186256 - Enhancer of terminal gene conversion, a new mutation in Drosophila melanogaster that induces telomere elongation by gene conversion.
Melnikova L, Georgiev P. Melnikova L, et al. Genetics. 2002 Nov;162(3):1301-12. doi: 10.1093/genetics/162.3.1301. Genetics. 2002. PMID: 12454074 Free PMC article. - Homolog-Dependent Repair Following Dicentric Chromosome Breakage in Drosophila melanogaster.
Bhandari J, Karg T, Golic KG. Bhandari J, et al. Genetics. 2019 Jul;212(3):615-630. doi: 10.1534/genetics.119.302247. Epub 2019 May 3. Genetics. 2019. PMID: 31053594 Free PMC article.
References
- Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989.
- Biessmann H, Mason J M, Ferry K, d'Hulst M, Valgeirsdottir K, Traverse K L, Pardue M L. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell. 1990;61:663–673. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases