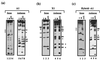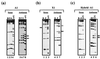Reprogrammable recognition codes in bicoid homeodomain-DNA interaction - PubMed (original) (raw)
Reprogrammable recognition codes in bicoid homeodomain-DNA interaction
V Dave et al. Mol Cell Biol. 2000 Oct.
Abstract
We describe experiments to determine how the homeodomain of the Drosophila morphogenetic protein Bicoid recognizes different types of DNA sequences found in natural enhancers. Our chemical footprint analyses reveal that the Bicoid homeodomain makes both shared and distinct contacts with a consensus site A1 (TAATCC) and a nonconsensus site X1 (TAAGCT). In particular, the guanine of X1 at position 4 (TAAGCT) is protected by Bicoid homeodomain. We provide further evidence suggesting that the unique arginine at position 54 (Arg 54) of the Bicoid homeodomain enables the protein to recognize X1 by specifically interacting with this position 4 guanine. We also describe experiments to analyze the contribution of artificially introduced Arg 54 to DNA recognition by other Bicoid-related homeodomains, including that from the human disease protein Pitx2. Our experiments demonstrate that the role of Arg 54 varies depending on the exact homeodomain framework and DNA sequences. Together, our results suggest that Bicoid and its related homeodomains utilize distinct recognition codes to interact with different DNA sequences, underscoring the need to study DNA recognition by Bicoid-class homeodomains in an individualized manner.
Figures
FIG. 1
A1 and X1 exhibit different methylation interference patterns for Bcd homeodomain binding. (a and b) A methylation interference analysis (see Materials and Methods for details) was performed using the Bcd homeodomain on both strands of DNA probes containing A1 (a) or X1 (b). (c) A third probe, Hybrid-A1, contains the recognition site A1 in the flanking sequences of X1; therefore, Hybrid-A1 and X1 probes are identical except for their recognition sequences. I and B represent the methylation profiles of DNA isolated from interfered (unbound) and bound fractions, respectively. In this assay, a band missing in the bound fraction indicates that methylation at this position interferes with (prevents) Bcd homeodomain binding. Strongly interfered guanine positions are marked with solid arrows, while partially interfered guanines are marked with open arrows. The interfered adenines are marked with open circles. The methylation interference data are summarized with the DNA sequences in Fig. 4. Two guanines on the antisense strand of X1 are highlighted with asterisks; these two positions exhibit strong methylation interference on X1 probe (b) but not on the Hybrid-A1 probe (c). A/G and G are Maxam-Gilbert DNA sequencing ladders.
FIG. 2
The Bcd homeodomain makes a conserved thymine contact on A1 and X1. DNA samples with one strand treated with KMnO4 (which modifies thymines) were used in binding assays to separate bound (B) and interfered (I; unbound) fractions. Thymine ladders (T) represent the positions of the modified thymines, used here as a reference. A band missing (marked by solid arrows in the bound fraction) indicates that modification of this thymine interferes with Bcd homeodomain binding. See the legend to Fig. 1 for further details.
FIG. 3
The Bcd homeodomain makes different guanine contacts on A1 and X1. A methylation protection analysis was performed on both strands of DNA probes containing A1 (a), X1 (b), or Hybrid-A1 (c). F and B represent the methylation profiles of DNA isolated from free (unbound) and bound fractions, respectively. In this assay, a band missing in the bound fraction indicates that the Bcd homeodomain protects this position from being methylated by DMS. See the legend to Fig. 1 for further details.
FIG. 4
Summary of interference and protection patterns on A1 and X1. Shown are the DMS interference and protection patterns on A1, X1, and Hybrid-A1. The solid and open arrows show guanines that are strongly and weakly interfered or protected, respectively. Open circles show adenine interference. Asterisks indicate interference at guanines on X1 that is not observed in Hybrid-A1. These two guanines are not protected by the Bcd homeodomain either (see the text for further details). The recognition sequences are in bold.
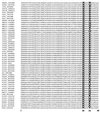
FIG. 5
Alignment of natural K50 homeodomains. Shown are sequences of known natural K50 homeodomains, highlighting amino acids 50 and 54. While alanine (A) and glutamine (Q) are the most frequently found residues at position 54, only the Bcd homeodomain contains an arginine at this position (Arg 54). The sequence data have been extracted from the Homeodomain Resource Database maintained by the Division of Intramural Research, Genome Technology Branch, National Human Genome Research Institute, National Institutes of Health (3). In this figure, the sequence of the Bcd homeodomain (HMBC_DROME) is listed in the first line.
FIG. 6
DNA recognition by the Bcd homeodomain and its derivatives. (a) Gel mobility shift assays using Bcd homeodomain and its mutants on three different DNA sites: A1 (TAATCC), X1 (TAAGCT), and X3s (TGATCC). See Materials and Methods for further details. The DNA probe concentration used in all these experiments was 10−9 M, and the estimated active-protein concentration was ∼10−10 M. (b) Dissociation constant (KD) measurements further confirm that the R54A mutation of the Bcd homeodomain preferentially affects its binding to X1. A Scatchard analysis was performed to determine the dissociation constants of the interactions between the Bcd homeodomain (either wild type or R54A mutant) and two DNA sequences (A1 and X1). For this assay, a quantitative gel shift analysis was performed at increasing concentrations of the DNA probes. The bound and free fractions of the probes were determined with a PhosphorImager and analyzed using Microsoft Excel (linear regression) to determine the KD value (−1/KD = slope of the plot of Bound/Free against Bound). The listed values represent three independent assays (mean ± standard deviation). For KD measurements, the wild-type (Wt) homeodomain was prepared in the same manner as the R54A mutant protein (see Materials and Methods). (c) Methylation protection by the Bcd(R54A) homeodomain on the sense strand of X1 (left), the antisense strand of X1 (middle), or the sense strand of another nonconsensus site X3s (right). A/G and G represent sequencing ladders, while B and F represent bound and free DNA, respectively. The sequence of the X3s sense strand probe is 5′-CTAGATCTGCTC
TGATCC
AGAATG-3′. The fourth-position guanine of the X1 sense strand that is protected by the wild-type Bcd homeodomain (Fig. 3b) is marked with an arrowhead. The guanines of X1 antisense and X3s sense strands that remain protected by the Bcd(R54A) homeodomain are marked with arrows.
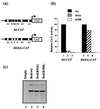
FIG. 7
Mutation of position 54 of the Bcd homeodomain reduces transcription activation in transient-transfection assays. (a) CAT reporter plasmids used in our transient-transfection assays in Drosophila Schneider S2 cells. hb-CAT contains a wild-type 250-bp enhancer element from hb, whereas hb(6A)-CAT contains a modified enhancer element with nonconsensus sites converted to consensus sites. (b) Relative CAT activities from four independent experiments, with wild-type (Wt) Bcd activity set at 100% for each reporter (lanes 1 and 4). Mutant Bcd(R54A) is defective in transcription activation from hb-CAT, while a modified hb enhancer containing all consensus A sites [_hb(6A)-CAT_] restores activation (compare lanes 2 and 5). Bcd(K50R), which contains a lysine-to-arginine change at position 50, does not show any activity from either reporters (lanes 3 and 6). The arbitrary activities are 100, 6.2, <1, 44.5, 35.4, and <1 for lanes 1 to 6, respectively. (c) Western blot analysis using HA-tagged Bcd derivatives. All the Bcd derivatives (lane 2, 3, and 4) accumulate at similar levels in the cell. Lane 1 represents cell lysate transfected with the vector expressing no activator.
FIG. 8
Effect of R54 on the DNA-binding activity of other homeodomains. (a to c) Gel mobility shift assay results obtained using different homeodomains and different DNA sites. The DNA probe concentration was 10−9 M for the experiments in panels a and c and 10−8 M for those in panel b. The active-protein concentration was normalized with respect to the Bcd homeodomain and kept at 10−9 M. See the text for further details. (d) Sequence alignment of the four natural K50-class homeodomains used in this study. The three helices of the homeodomains are marked. Wt, wild type.
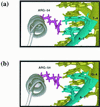
FIG. 9
Adaptive interaction of the Bcd homeodomain on consensus (A1) and nonconsensus (X1) sites mediated by Arg 54. Shown are homology-modeled structures of the Bcd homeodomain complexed with site A1 (a) or X1 (b). Only the third helix (pointing toward the page) and the side chain of Arg 54 are shown in this figure for clarity. For A1, N-7 of the third-base adenine (TA
A
TCC; the corresponding base pair is colored in cyan) is in the vicinity of the NH2 group emanating from Arg 54 (colored in magenta) and may form a single hydrogen bond (shown in yellow). For the fourth-position thymine, the NH2 group of Arg 54 is too far away to form a hydrogen bond (>3 Å). For X1, the side chain of Arg 54 swings upward with a vertical translation, allowing the NH2 groups to move about 1.4 Å in order to form a second hydrogen bond with the fourth-position guanine (TAA
G
CC). Thus, Arg 54 forms bidentate hydrogen bonds on X1 in this model: one with N-7 of the third-base adenine and the other with O-6 of the fourth base guanine (TA
AG
CC; the corresponding base pairs are colored in cyan). In this model structure, as seen with other homeodomains (72), Asn 51 (not shown in the figure) is well positioned to contact the third-base adenine, and therefore we propose that the N-7 position of this adenine engages in shared hydrogen bonds: one from Asn 51 and the other from Arg 54.
Similar articles
- The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
Baird-Titus JM, Clark-Baldwin K, Dave V, Caperelli CA, Ma J, Rance M. Baird-Titus JM, et al. J Mol Biol. 2006 Mar 10;356(5):1137-51. doi: 10.1016/j.jmb.2005.12.007. Epub 2005 Dec 22. J Mol Biol. 2006. PMID: 16406070 - Isolation of mutations that disrupt cooperative DNA binding by the Drosophila bicoid protein.
Burz DS, Hanes SD. Burz DS, et al. J Mol Biol. 2001 Jan 12;305(2):219-30. doi: 10.1006/jmbi.2000.4287. J Mol Biol. 2001. PMID: 11124901 - Target selectivity of bicoid is dependent on nonconsensus site recognition and protein-protein interaction.
Zhao C, Dave V, Yang F, Scarborough T, Ma J. Zhao C, et al. Mol Cell Biol. 2000 Nov;20(21):8112-23. doi: 10.1128/MCB.20.21.8112-8123.2000. Mol Cell Biol. 2000. PMID: 11027281 Free PMC article. - Drosophila development: homeodomains and translational control.
Zamore PD, Lehmann R. Zamore PD, et al. Curr Biol. 1996 Jul 1;6(7):773-5. doi: 10.1016/s0960-9822(02)00591-2. Curr Biol. 1996. PMID: 8805294 Review. - Homeodomain interactions.
Wolberger C. Wolberger C. Curr Opin Struct Biol. 1996 Feb;6(1):62-8. doi: 10.1016/s0959-440x(96)80096-0. Curr Opin Struct Biol. 1996. PMID: 8696974 Review.
Cited by
- Combinatorial interactions of two amino acids with a single base pair define target site specificity in plant dimeric homeodomain proteins.
Tron AE, Bertoncini CW, Palena CM, Chan RL, Gonzalez DH. Tron AE, et al. Nucleic Acids Res. 2001 Dec 1;29(23):4866-72. doi: 10.1093/nar/29.23.4866. Nucleic Acids Res. 2001. PMID: 11726696 Free PMC article. - Pitx2a expression alters actin-myosin cytoskeleton and migration of HeLa cells through Rho GTPase signaling.
Wei Q, Adelstein RS. Wei Q, et al. Mol Biol Cell. 2002 Feb;13(2):683-97. doi: 10.1091/mbc.01-07-0358. Mol Biol Cell. 2002. PMID: 11854422 Free PMC article. - Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites.
Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Noyes MB, et al. Cell. 2008 Jun 27;133(7):1277-89. doi: 10.1016/j.cell.2008.05.023. Cell. 2008. PMID: 18585360 Free PMC article. - Identification of a novel frameshift mutation in PITX2 gene in a Chinese family with Axenfeld-Rieger syndrome.
Yin HF, Fang XY, Jin CF, Yin JF, Li JY, Zhao SJ, Miao Q, Song FW. Yin HF, et al. J Zhejiang Univ Sci B. 2014 Jan;15(1):43-50. doi: 10.1631/jzus.B1300053. J Zhejiang Univ Sci B. 2014. PMID: 24390743 Free PMC article. - A single Hox3 gene with composite bicoid and zerknullt expression characteristics in non-Cyclorrhaphan flies.
Stauber M, Prell A, Schmidt-Ott U. Stauber M, et al. Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):274-9. doi: 10.1073/pnas.012292899. Epub 2002 Jan 2. Proc Natl Acad Sci U S A. 2002. PMID: 11773616 Free PMC article.
References
- Akman S A, Doroshow J H, Dizdaroglu M. Base modifications in plasmid DNA caused by potassium permanganate. Arch Biochem Biophys. 1990;282:202–205. - PubMed
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994.
- Billeter M, Qian Y Q, Otting G, Muller M, Gehring W, Wuthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993;234:1084–1093. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases