CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation - PubMed (original) (raw)
CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation
S B Hake et al. Mol Cell Biol. 2000 Oct.
Abstract
The major histocompatibility complex (MHC) class II transactivator CIITA plays a pivotal role in the control of the cellular immune response through the quantitative regulation of MHC class II expression. We have analyzed a region of CIITA with similarity to leucine-rich repeats (LRRs). CIITA LRR alanine mutations abolish both the transactivation capacity of full-length CIITA and the dominant-negative phenotype of CIITA mutants with N-terminal deletions. We demonstrate direct interaction of CIITA with the MHC class II promoter binding protein RFX5 and could also detect novel interactions with RFXANK, NF-YB, and -YC. However, none of these interactions is influenced by CIITA LRR mutagenesis. On the other hand, chromatin immunoprecipitation shows that in vivo binding of CIITA to the MHC class II promoter is dependent on LRR integrity. LRR mutations lead to an impaired nuclear localization of CIITA, indicating that a major function of the CIITA LRRs is in nucleocytoplasmic translocation. There is, however, evidence that the CIITA LRRs are also involved more directly in MHC class II gene transactivation. CIITA interacts with a novel protein of 33 kDa in a manner sensitive to LRR mutagenesis. CIITA is therefore imported into the nucleus by an LRR-dependent mechanism, where it activates transcription through multiple protein-protein interactions with the MHC class II promoter binding complex.
Figures
FIG. 1
Generation of CIITA LRR alanine mutants. (A) Alignment of RI with the LRRs of CIITA. The alternating A- and B-type repeats of porcine RI (accession no. P10775) are aligned separately on the upper left and right sides. Amino acids that are in contact with the substrate RNase A are indicated as follows: those involved in van der Waals interactions are shaded, and those participating in hydrogen bond formation are in lowercase (25). The consensus sequences of the RI repeats are shown below the RI sequence, and positions participating in substrate binding are shaded. The LRR regions of human CIITA (amino acid positions 988 to 1097) are at the bottom. Potential contact positions were combined from both types of RI repeats, and the corresponding positions in CIITA are shaded. (B) CIITA LRR multiple-alanine mutants. At the top the RI B-type repeat consensus (cons.) sequence is shown. Compiled contact positions are indicated together with the delimitation of β-strand-, loop-, and α-helical regions. The positions of the multiple alanine mutations are indicated by circles. The names of the mutants are indicated on the left. Open circles, mutations without effect on CIITA function; solid circles, mutations leading to a loss of CIITA transactivation activity (see Fig. 2A). (C) CIITA LRR single-alanine mutants. Solid circles, mutations which abolish CIITA function in single-alanine mutants (see Fig. 2B); open circles, mutations without effect in single-alanine mutants. The single alanine mutations are identified by their amino acid positions.
FIG. 2
FACS analysis of CIITA LRR alanine mutants. (A) Multiple-alanine mutants. Wild-type and mutant CIITA cDNAs in the episomal expression vector EBS-PL were transfected into the CIITA-deficient cell line RJ2.2.5 and analyzed in bulk for HLA-DR expression by FACS after 2 weeks of antibiotic selection. The mutants are presented in N-terminal-to-C-terminal order (see also Fig. 1B). (B) Single-alanine mutants. All seven mutants with mutations which abolish CIITA function and two mutants with mutations without effect are shown. Mutations are identified by their amino acid positions (see also Fig. 1C). (C) Protein expression analysis of CIITA LRR multiple-alanine mutants. CIITA cDNA expression constructs were transfected into HEK293-EBNA cells, and transient protein expression was analyzed by Western blotting with a CIITA-specific antiserum. The blot shows two independent transfections for each construct. The two major bands of CIITA correspond most probably to products resulting from initiation at the first and second AUGs of the CIITA cDNA, form III (32, 45), used here. (D) Protein expression analysis of CIITA LRR single-alanine mutants. Only the mutants that have lost their transactivation potential are identified individually; the others are indicated by a horizontal bar.
FIG. 2
FACS analysis of CIITA LRR alanine mutants. (A) Multiple-alanine mutants. Wild-type and mutant CIITA cDNAs in the episomal expression vector EBS-PL were transfected into the CIITA-deficient cell line RJ2.2.5 and analyzed in bulk for HLA-DR expression by FACS after 2 weeks of antibiotic selection. The mutants are presented in N-terminal-to-C-terminal order (see also Fig. 1B). (B) Single-alanine mutants. All seven mutants with mutations which abolish CIITA function and two mutants with mutations without effect are shown. Mutations are identified by their amino acid positions (see also Fig. 1C). (C) Protein expression analysis of CIITA LRR multiple-alanine mutants. CIITA cDNA expression constructs were transfected into HEK293-EBNA cells, and transient protein expression was analyzed by Western blotting with a CIITA-specific antiserum. The blot shows two independent transfections for each construct. The two major bands of CIITA correspond most probably to products resulting from initiation at the first and second AUGs of the CIITA cDNA, form III (32, 45), used here. (D) Protein expression analysis of CIITA LRR single-alanine mutants. Only the mutants that have lost their transactivation potential are identified individually; the others are indicated by a horizontal bar.
FIG. 2
FACS analysis of CIITA LRR alanine mutants. (A) Multiple-alanine mutants. Wild-type and mutant CIITA cDNAs in the episomal expression vector EBS-PL were transfected into the CIITA-deficient cell line RJ2.2.5 and analyzed in bulk for HLA-DR expression by FACS after 2 weeks of antibiotic selection. The mutants are presented in N-terminal-to-C-terminal order (see also Fig. 1B). (B) Single-alanine mutants. All seven mutants with mutations which abolish CIITA function and two mutants with mutations without effect are shown. Mutations are identified by their amino acid positions (see also Fig. 1C). (C) Protein expression analysis of CIITA LRR multiple-alanine mutants. CIITA cDNA expression constructs were transfected into HEK293-EBNA cells, and transient protein expression was analyzed by Western blotting with a CIITA-specific antiserum. The blot shows two independent transfections for each construct. The two major bands of CIITA correspond most probably to products resulting from initiation at the first and second AUGs of the CIITA cDNA, form III (32, 45), used here. (D) Protein expression analysis of CIITA LRR single-alanine mutants. Only the mutants that have lost their transactivation potential are identified individually; the others are indicated by a horizontal bar.
FIG. 3
Coimmunoprecipitation of CIITA and MHC-II promoter binding proteins. (A) CIITA and RFX5. EGFP-CIITA and Myc-RFX5 were transfected individually or together into HEK293-EBNA cells and immunoprecipitated with an anti-GFP serum. Coprecipitated RFX5 was detected with a myc antibody in a Western blot analysis. Lane 6, cotransfection of CIITA and RFX5; lanes 1 to 5, controls. All CIITA multiple-alanine mutants coprecipitated RFX5 (lanes 7 and 8; data not shown). Lanes 10 and 11, Western blot controls of the material used in lanes 5 and 6, respectively; lane 9, molecular weight standard. (B) CIITA and RFXANK. Interaction between EGFP-CIITA and RFXANK was analyzed as for RFX5 with the exception that unmodified RFXANK was transfected and that an RFXANK-specific polyclonal antiserum was used for detection. Again, all CIITA multiple-alanine mutants coimmunoprecipitated RFXANK (lanes 7 and 8; data not shown). The band detected in the EGFP-CIITA Western blot control (lane 10) migrates below that of RFXANK and is therefore nonspecific. (C and D) CIITA and NF-YB and NF-YC. Immunoprecipitations were carried out here with a GFP-specific serum and Dynabeads. The calculated molecular masses for the various constructs are as follows: Myc6-RFX5, 62 kDa; RFXANK, 29 kDa; Myc6-NF-YB, 33 kDa; Myc6-NF-YC, 47 kDa. Arrowheads, positions of bands of the expected sizes.
FIG. 4
In vivo recruitment of CIITA to the MHC-II promoter. (A) CIITA LRR mutations abolish dominant-negative effect. Plasmids encoding CIITA LRR mutants were cotransfected with equal amounts of wild-type CIITA plasmid in HEK293-EBNA cells, and HLA-DQ expression was analyzed 72 h later. (Top) Open profile, results for cells transfected with empty vector alone; shaded profiles, cotransfection of CIITA wild-type cDNA either with empty vector (left; this profile is used as an overlay for comparison in the middle and bottom portions and is shown there as a solid line), a weak dominant-negative CIITA mutant, NLS-102 (middle), and a strongly dominant-negative mutant, NLS-L335 (right; this profile is shown in an overlay for comparison in the middle and lower portions as a dashed line). (Middle) CIITA-MT5, -MT7, and -MT10 (shaded profiles) were cotransfected with wild-type CIITA and analyzed as described above. (Bottom) The LRR alanine mutations of MT5, MT7, and MT10 (shaded profiles) were introduced into the N-terminal deletion mutant NLS-L335 and analyzed as described above. (B) CIITA LRR mutations impair the in vivo recruitment of CIITA to the HLA-DRA promoter. Transient transfectants of wild-type and LRR-mutated EGFP-CIITA constructs in HEK293-EBNA cells were cross-linked in vivo with formaldehyde, and cross-linked protein-chromatin complexes were immunoprecipitated with GFP-specific monoclonal antibodies. Precipitation of the HLA-DRA promoter was analyzed by PCR amplification.
FIG. 5
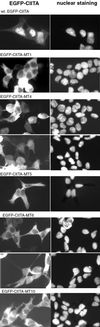
Subcellular localization of CIITA LRR mutants. HEK293-EBNA cells were transfected with wild-type (wt) or LRR-mutated EGFP-CIITA constructs as indicated and analyzed 48 h later. Left, GFP fluorescence; right, nuclear staining with bis-benzimidine. The lower images of EGFP-MT4 and -MT6 show cells with a very low level of GFP fluorescence.
FIG. 6
Immunoprecipitation of in vivo-labeled CIITA transfectants. Wild-type EGFP-CIITA and EGFP-CIITA LRR multiple-alanine mutants were transfected individually into HEK293-EBNA cells, and the cells were metabolically labeled overnight with [35S]methionine. EGFP-CIITA and associated proteins were immunoprecipitated with a GFP-specific polyclonal antiserum (Clontech), followed by SDS-PAGE on 7.5 or 15% gels and autoradiography. (A) A 15% gel of cells transfected with EGFP, EGFP-CIITA, and all nine EGFP-CIITA multiple-alanine mutants that abolish MHC-II transactivation. (B) Analysis of mutants that have retained their transactivation potential. Arrowheads, positions of EGFP, the CIITA-associated 33-kDa band, and EGFP-CIITA. Two of the mutants, MT2 and MT11, became unstable when fused to EGFP (lanes 4 and 9).
FIG. 7
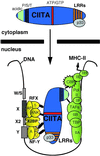
LRR-dependent nuclear localization of CIITA and MHC-II transactivation through multiple protein-protein interactions. CBP, CREB binding protein; TBP, TATA-binding protein; TAFs, TBP-associated factors.
Similar articles
- Importance of class II transactivator leucine-rich repeats for dominant-negative function and nucleo-cytoplasmic transport.
Camacho-Carvajal MM, Klingler S, Schnappauf F, Hake SB, Steimle V. Camacho-Carvajal MM, et al. Int Immunol. 2004 Jan;16(1):65-75. doi: 10.1093/intimm/dxh010. Int Immunol. 2004. PMID: 14688062 - Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation.
Zika E, Greer SF, Zhu XS, Ting JP. Zika E, et al. Mol Cell Biol. 2003 May;23(9):3091-102. doi: 10.1128/MCB.23.9.3091-3102.2003. Mol Cell Biol. 2003. PMID: 12697811 Free PMC article. - Expression of MHC II genes.
Drozina G, Kohoutek J, Jabrane-Ferrat N, Peterlin BM. Drozina G, et al. Curr Top Microbiol Immunol. 2005;290:147-70. doi: 10.1007/3-540-26363-2_7. Curr Top Microbiol Immunol. 2005. PMID: 16480042 Review. - The MHC Class II Transactivator CIITA: Not (Quite) the Odd-One-Out Anymore among NLR Proteins.
León Machado JA, Steimle V. León Machado JA, et al. Int J Mol Sci. 2021 Jan 22;22(3):1074. doi: 10.3390/ijms22031074. Int J Mol Sci. 2021. PMID: 33499042 Free PMC article. Review.
Cited by
- Phosphorylation of CIITA directs its oligomerization, accumulation and increased activity on MHCII promoters.
Tosi G, Jabrane-Ferrat N, Peterlin BM. Tosi G, et al. EMBO J. 2002 Oct 15;21(20):5467-76. doi: 10.1093/emboj/cdf557. EMBO J. 2002. PMID: 12374747 Free PMC article. - Promoter-specific functions of CIITA and the MHC class II enhanceosome in transcriptional activation.
Masternak K, Reith W. Masternak K, et al. EMBO J. 2002 Mar 15;21(6):1379-88. doi: 10.1093/emboj/21.6.1379. EMBO J. 2002. PMID: 11889043 Free PMC article. - NLRP2 is a suppressor of NF-ƙB signaling and HLA-C expression in human trophoblasts†,‡.
Tilburgs T, Meissner TB, Ferreira LMR, Mulder A, Musunuru K, Ye J, Strominger JL. Tilburgs T, et al. Biol Reprod. 2017 Apr 1;96(4):831-842. doi: 10.1093/biolre/iox009. Biol Reprod. 2017. PMID: 28340094 Free PMC article. - Epigenetic regulation of major histocompatibility complexes in gastrointestinal malignancies and the potential for clinical interception.
Tovar Perez JE, Zhang S, Hodgeman W, Kapoor S, Rajendran P, Kobayashi KS, Dashwood RH. Tovar Perez JE, et al. Clin Epigenetics. 2024 Jun 24;16(1):83. doi: 10.1186/s13148-024-01698-8. Clin Epigenetics. 2024. PMID: 38915093 Free PMC article. Review. - Expression of the three human major histocompatibility complex class II isotypes exhibits a differential dependence on the transcription factor RFXAP.
Peretti M, Villard J, Barras E, Zufferey M, Reith W. Peretti M, et al. Mol Cell Biol. 2001 Sep;21(17):5699-709. doi: 10.1128/MCB.21.17.5699-5709.2001. Mol Cell Biol. 2001. PMID: 11486010 Free PMC article.
References
- Becker J, Melchior F, Gerke V, Bischoff F R, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. - PubMed
- Bogan A A, Thorn K S. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. - PubMed
- Bontron S, Steimle V, Ucla C, Eibl M M, Mach B. Two novel mutations in the MHC class II transactivator CIITA in a second patient from MHC class II deficiency complementation group A. Hum Genet. 1997;99:541–546. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials