PCR primers that amplify fungal rRNA genes from environmental samples - PubMed (original) (raw)
PCR primers that amplify fungal rRNA genes from environmental samples
J Borneman et al. Appl Environ Microbiol. 2000 Oct.
Abstract
Two PCR primer pairs were designed to amplify rRNA genes (rDNA) from all four major phyla of fungi: Ascomycota, Basidiomycota, Chytridomycota, and Zygomycota. PCRs performed with these primers showed that both pairs amplify DNA from organisms representing the major taxonomic groups of fungi but not from nonfungal sources. To test the ability of the primers to amplify fungal rDNA from environment samples, clone libraries from two avocado grove soils were constructed and analyzed. These soils possess different abilities to inhibit avocado root rot caused by Phythophthora cinnamomi. Analysis of the two rDNA clone libraries revealed differences in the two fungal communities. It also revealed a markedly different depiction of the soil fungal community than that generated by a culture-based analysis, confirming the value of rDNA-based approaches for identifying organisms that may not readily grow on agar media. Additional evidence of the usefulness of the primers was obtained by identifying fungi associated with avocado leaves. In both the soil and leaf analyses, no nonfungal rDNA sequences were identified, illustrating the selectivity of these PCR primers. This work demonstrates the ability of two newly developed PCR primer sets to amplify fungal rDNA from soil and plant tissue, thereby providing unique tools to examine this vast and mostly undescribed community of organisms.
Figures
FIG. 1
Diagram of the eukaryotic small-subunit rDNA with the variable regions highlighted in gray. The numerical positions of the primers and the PCR product sizes were obtained by using S. cereviseae (GenBank accession no. J01353) as the reference template.
FIG. 2
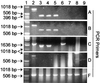
PCR amplification of representative DNA templates with five different fungal rDNA primer sets. PCR products were resolved on agarose gels and stained with ethidium bromide. (A) Primers nu-SSU-0817-5′ and nu-SSU-1196-3′; (B) primers nu-SSU-0817-5′ and nu-SSU-1536-3′; (C) primers EF4 and EF3; (D) primers EF4 and NS3; (E) primers EF4 and fung5. Lane 1, 1-kb ladder (Gibco BRL, Grand Island, N.Y.); lane 2, Monilinia fructicola; lane 3, Tilletia caries; lane 4, Allomyces javanicus; lane 5, Glomus deserticola; lane 6, Escherichia coli; lane 7, Caenorhabditis elegans; lane 8, Cucumis melo; lane 9, Phytophthora infestans. The larger sizes of the M. fructicola bands (lane 2) are likely due to intron insertion. The results of these experiments and a more extensive analysis are summarized in Table 2.
FIG. 3
PCR amplification of DNA extracted from two soils with five different fungal rDNA primer pairs. PCR products were resolved on agarose gels and stained with ethidium bromide. Lane 1, primers EF4 and EF3; lane 2, primers EF4 and fung5; lane 3, primers EF4 and NS3; lane 4, primers nu-SSU-0817-5′ and nu-SSU-1196-3′; lanes 5 and 6, primers nu-SSU-0817-5′ and nu-SSU-1536-3′; lanes 1 through 5, Vanoni soil DNA; lane 6, Powell soil DNA; lane 7, 1-kb ladder (Gibco BRL).
References
- Apinis A E. Facts and problems. Mycopathol Mycol Appl. 1972;48:93–109. - PubMed
- Bock M, Maiwald M, Kappe R, Nickel P, Naeher H. Polymerase chain reaction-based detection of dermatophyte DNA with a fungus-specific primer system. Mycoses. 1994;37:79–84. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases