Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins - PubMed (original) (raw)
Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins
A Rak et al. EMBO J. 2000.
Abstract
We present the 1.9 A resolution crystal structure of the catalytic domain of Gyp1p, a specific GTPase activating protein (GAP) for Ypt proteins, the yeast homologues of Rab proteins, which are involved in vesicular transport. Gyp1p is a member of a large family of eukaryotic proteins with shared sequence motifs. Previously, no structural information was available for any member of this class of proteins. The GAP domain of Gyp1p was found to be fully alpha-helical. However, the observed fold does not superimpose with other alpha-helical GAPs (e.g. Ras- and Cdc42/Rho-GAP). The conserved and catalytically crucial arginine residue, identified by mutational analysis, is in a comparable position to the arginine finger in the Ras- and Cdc42-GAPs, suggesting that Gyp1p utilizes an arginine finger in the GAP reaction, in analogy to Ras- and Cdc42-GAPs. A model for the interaction between Gyp1p and the Ypt protein satisfying biochemical data is given.
Figures
Fig. 1. Three-dimensional structure of the Ypt-GAP domain of Gyp1p. The ribbon diagram displays the secondary structure elements and the active site arginine residue in ball-and-stick representation. Regions of the seven shared sequence motifs A–F are highlighted in green. This figure was generated using the programs BOBSCRIPT (Esnouf, 1997) and raster3D (Merritt and Murphy, 1994).
Fig. 2. Sequence comparison of the catalytically active domains of Gyp1p homologs. The sequence alignment was performed with the program CLUSTAL_W (Thompson et al., 1994) and adjusted manually in the loop regions. Only the residues modelled in the crystal structure of Gyp1-46p were included in the alignment. The residue numbering is based on the Gyp1p full-length sequence. Amino acid residues strictly conserved have a red background, residues (hydrophobic, aromatic, polar, negatively or positively charged) conserved within four or more Gyp proteins are indicated with a yellow background. Secondary structure elements are shown above the aligned sequences. The six shared motifs named A–F, found previously to be common between Ypt-GAPs and proteins involved in spindle checkpoint assembly (Neuwald, 1997), are underlined with solid bars in green. The blue stars indicate residues that are at the surface of the putative active site cleft of Gyp1-46p. The red line indicates the region from residue 510–569 which is not included in the crystal structure. The number for the first and last residue used for the alignment is given together with the overall length of the Gyp protein in parenthesis if the last residue is not the C-terminus. The figure was prepared with the program ESPript (Gouet et al., 1999).
Fig. 3. Central core and active site of the Ypt-GAP domain of Gyp1p. (A) Stereo view of the final 2_F_o – _F_c electron density map contoured at the 2σ level displaying the aromatic core of Gyp1-46p. Displayed are the highly conserved Trp290 of motif A and the surrounding hydrophobic residues. (B) Stereo view of the final 2_F_o – _F_c electron density map contoured at the 2σ level at the active site region of Gyp1-46p. The refined model (ball-and-stick) is superimposed, displaying the residues 339–344 of motif B which includes the highly conserved Asp340 and the catalytically active residue Arg343. This figure was generated using the programs BOBSCRIPT (Esnouf, 1997) and raster3D (Merritt and Murphy, 1994).
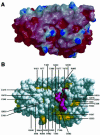
Fig. 4. The putative Ypt binding cleft. (A) Electrostatic surface representations viewed into the concave side of the molecule. The figure was generated using the program GRASP (Nicholls et al., 1993) and rendered with the program raster3D (Merritt and Murphy, 1994). Red indicates negatively charged (–7 kT) and blue positively charged regions (+7 kT). (B) Surface CPK representation of the Gyp1p Ypt-GAP domain shown in the same orientation as in (A). Residues that are highly conserved in the different Gyp Ypt-GAP domains are coloured pink; well conserved residues are coloured yellow (see sequence alignment, Figure 2). Residues that are solvent accessible and form the surface of the cleft are labelled.
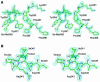
Fig. 5. Docking approach for the complex between Gyp1p and Ypt51p-GTP, modelled manually on the basis of known GAP–GTPase structures. (A) Active site of the complex formed between p120-GAP and H-Ras p21(GDP-AlF3). The hydrogen bonds between GAP residues Arg789 and Arg903 and H-Ras p21 residues Gln61 and Glu63 as well as with AlF3 are indicated. This specific hydrogen bond network and the orientation of the side chains were used as a model for manual docking of Ypt51-GTP to Gyp1-46p. (B) Close-up view of the active site in the putative Gyp1-46p–Ypt51-GTP complex. For the interaction between the side chain of Arg343 and the γ-phosphate group, the salt bridge formed between Arg343 and Asp340 has to be broken. Gln66 of Ypt51p is well oriented to become positioned closer to the γ-phosphate by forming a hydrogen bond between its side chain and the main chain carbonyl group of Arg343 of Gyp1p. (C) Ribbon representation of the putative complex. The orientation of Gyp1-46p is the same as in Figure 1. The essential arginine Arg343 of Gyp1p, the active site glutamine Gln66 of Ypt51p and the bound nucleotide GTP are shown in ball-and-stick representation. This figure was generated using the programs BOBSCRIPT (Esnouf, 1997) and raster3D (Merritt and Murphy, 1994).
Similar articles
- TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism.
Pan X, Eathiraj S, Munson M, Lambright DG. Pan X, et al. Nature. 2006 Jul 20;442(7100):303-6. doi: 10.1038/nature04847. Nature. 2006. PMID: 16855591 - Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP.
Albert S, Gallwitz D. Albert S, et al. Biol Chem. 2000 May-Jun;381(5-6):453-6. doi: 10.1515/BC.2000.059. Biol Chem. 2000. PMID: 10937877 - TBC proteins: GAPs for mammalian small GTPase Rab?
Fukuda M. Fukuda M. Biosci Rep. 2011 Jun;31(3):159-68. doi: 10.1042/BSR20100112. Biosci Rep. 2011. PMID: 21250943 Review. - GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila.
Bernards A. Bernards A. Biochim Biophys Acta. 2003 Mar 17;1603(2):47-82. doi: 10.1016/s0304-419x(02)00082-3. Biochim Biophys Acta. 2003. PMID: 12618308 Review.
Cited by
- The TRE17 oncogene encodes a component of a novel effector pathway for Rho GTPases Cdc42 and Rac1 and stimulates actin remodeling.
Masuda-Robens JM, Kutney SN, Qi H, Chou MM. Masuda-Robens JM, et al. Mol Cell Biol. 2003 Mar;23(6):2151-61. doi: 10.1128/MCB.23.6.2151-2161.2003. Mol Cell Biol. 2003. PMID: 12612085 Free PMC article. - A fragment activity assay reveals the key residues of TBC1D15 GTPase-activating protein (GAP) in Chiloscyllium plagiosum.
Jin Y, Lin G, Chen Y, Ge Y, Liang R, Wu J, Chen J, Wang D, Shi H, Fei H, Lv Z. Jin Y, et al. BMC Mol Biol. 2019 Feb 12;20(1):5. doi: 10.1186/s12867-019-0122-2. BMC Mol Biol. 2019. PMID: 30755162 Free PMC article. - Arf1 orchestrates Rab GTPase conversion at the _trans_-Golgi network.
Thomas LL, Highland CM, Fromme JC. Thomas LL, et al. Mol Biol Cell. 2021 May 15;32(11):1104-1120. doi: 10.1091/mbc.E20-10-0664. Epub 2021 Mar 31. Mol Biol Cell. 2021. PMID: 33788577 Free PMC article. - N-terminal tyrosine residues within the potassium channel Kir3 modulate GTPase activity of Galphai.
Ippolito DL, Temkin PA, Rogalski SL, Chavkin C. Ippolito DL, et al. J Biol Chem. 2002 Sep 6;277(36):32692-6. doi: 10.1074/jbc.M204407200. Epub 2002 Jun 24. J Biol Chem. 2002. PMID: 12082117 Free PMC article. - Crystal structures of human TBC1D1 and TBC1D4 (AS160) RabGTPase-activating protein (RabGAP) domains reveal critical elements for GLUT4 translocation.
Park SY, Jin W, Woo JR, Shoelson SE. Park SY, et al. J Biol Chem. 2011 May 20;286(20):18130-8. doi: 10.1074/jbc.M110.217323. Epub 2011 Mar 23. J Biol Chem. 2011. PMID: 21454505 Free PMC article.
References
- Ahmadian M.R., Stege,P., Scheffzek,K. and Wittinghofer,A. (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nature Struct. Biol., 4, 686–689. - PubMed
- Albert S. and Gallwitz,D. (1999) Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J. Biol. Chem., 274, 33186–33189. - PubMed
- Albert S. and Gallwitz,D. (2000) Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol. Chem., 381, 453–456. - PubMed
- Bernstein F.C., Koetzle,T.F., Williams,G.J.B., Meyer,E.T.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The protein data bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous