16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates - PubMed (original) (raw)
16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates
M Drancourt et al. J Clin Microbiol. 2000 Oct.
Abstract
Some bacteria are difficult to identify with phenotypic identification schemes commonly used outside reference laboratories. 16S ribosomal DNA (rDNA)-based identification of bacteria potentially offers a useful alternative when phenotypic characterization methods fail. However, as yet, the usefulness of 16S rDNA sequence analysis in the identification of conventionally unidentifiable isolates has not been evaluated with a large collection of isolates. In this study, we evaluated the utility of 16S rDNA sequencing as a means to identify a collection of 177 such isolates obtained from environmental, veterinary, and clinical sources. For 159 isolates (89.8%) there was at least one sequence in GenBank that yielded a similarity score of > or =97%, and for 139 isolates (78.5%) there was at least one sequence in GenBank that yielded a similarity score of > or =99%. These similarity score values were used to defined identification at the genus and species levels, respectively. For isolates identified to the species level, conventional identification failed to produce accurate results because of inappropriate biochemical profile determination in 76 isolates (58.7%), Gram staining in 16 isolates (11.6%), oxidase and catalase activity determination in 5 isolates (3.6%) and growth requirement determination in 2 isolates (1.5%). Eighteen isolates (10.2%) remained unidentifiable by 16S rDNA sequence analysis but were probably prototype isolates of new species. These isolates originated mainly from environmental sources (P = 0.07). The 16S rDNA approach failed to identify Enterobacter and Pantoea isolates to the species level (P = 0.04; odds ratio = 0.32 [95% confidence interval, 0.10 to 1.14]). Elsewhere, the usefulness of 16S rDNA sequencing was compromised by the presence of 16S rDNA sequences with >1% undetermined positions in the databases. Unlike phenotypic identification, which can be modified by the variability of expression of characters, 16S rDNA sequencing provides unambiguous data even for rare isolates, which are reproducible in and between laboratories. The increase in accurate new 16S rDNA sequences and the development of alternative genes for molecular identification of certain taxa should further improve the usefulness of molecular identification of bacteria.
Figures
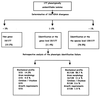
FIG. 1
Identification scheme for 177 phenotypically unidentifiable bacterial isolates.
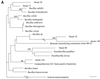
FIG. 2
Distance-related trees indicating the phylogenetic relationships of 18 unidentified isolates referred as strain numbers. (A) Low-percent-G+C-content gram-positive isolates; (B) high-percent-G+C-content gram-positive isolates; (C) gamma subgroup Proteobacteria isolates; (D) Bacteroides-Cytophaga subgroup isolates. The numbers at nodes are the proportions of 100 bootstrap resamplings that support the topology shown. Only bootstrap values of >90% are indicated. Bars, 1% divergence.
FIG. 2
Distance-related trees indicating the phylogenetic relationships of 18 unidentified isolates referred as strain numbers. (A) Low-percent-G+C-content gram-positive isolates; (B) high-percent-G+C-content gram-positive isolates; (C) gamma subgroup Proteobacteria isolates; (D) Bacteroides-Cytophaga subgroup isolates. The numbers at nodes are the proportions of 100 bootstrap resamplings that support the topology shown. Only bootstrap values of >90% are indicated. Bars, 1% divergence.
FIG. 2
Distance-related trees indicating the phylogenetic relationships of 18 unidentified isolates referred as strain numbers. (A) Low-percent-G+C-content gram-positive isolates; (B) high-percent-G+C-content gram-positive isolates; (C) gamma subgroup Proteobacteria isolates; (D) Bacteroides-Cytophaga subgroup isolates. The numbers at nodes are the proportions of 100 bootstrap resamplings that support the topology shown. Only bootstrap values of >90% are indicated. Bars, 1% divergence.
FIG. 2
Distance-related trees indicating the phylogenetic relationships of 18 unidentified isolates referred as strain numbers. (A) Low-percent-G+C-content gram-positive isolates; (B) high-percent-G+C-content gram-positive isolates; (C) gamma subgroup Proteobacteria isolates; (D) Bacteroides-Cytophaga subgroup isolates. The numbers at nodes are the proportions of 100 bootstrap resamplings that support the topology shown. Only bootstrap values of >90% are indicated. Bars, 1% divergence.
Similar articles
- 16S rRNA sequencing in routine bacterial identification: a 30-month experiment.
Mignard S, Flandrois JP. Mignard S, et al. J Microbiol Methods. 2006 Dec;67(3):574-81. doi: 10.1016/j.mimet.2006.05.009. Epub 2006 Jul 21. J Microbiol Methods. 2006. PMID: 16859787 - Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles.
Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, Leung KW, Tam DM, Que TL, Yuen KY. Woo PC, et al. J Clin Microbiol. 2003 May;41(5):1996-2001. doi: 10.1128/JCM.41.5.1996-2001.2003. J Clin Microbiol. 2003. PMID: 12734240 Free PMC article. - Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories.
Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Woo PC, et al. Clin Microbiol Infect. 2008 Oct;14(10):908-34. doi: 10.1111/j.1469-0691.2008.02070.x. Clin Microbiol Infect. 2008. PMID: 18828852 Review. - Utility of 16S rDNA Sequencing for Identification of Rare Pathogenic Bacteria.
Loong SK, Khor CS, Jafar FL, AbuBakar S. Loong SK, et al. J Clin Lab Anal. 2016 Nov;30(6):1056-1060. doi: 10.1002/jcla.21980. Epub 2016 May 17. J Clin Lab Anal. 2016. PMID: 27184222 Free PMC article. - Thinking beside the box: Should we care about the non-coding strand of the 16S rRNA gene?
Garcia-Mazcorro JF, Barcenas-Walls JR. Garcia-Mazcorro JF, et al. FEMS Microbiol Lett. 2016 Aug;363(16):fnw171. doi: 10.1093/femsle/fnw171. Epub 2016 Jul 12. FEMS Microbiol Lett. 2016. PMID: 27412167 Review.
Cited by
- Desert gerbils affect bacterial composition of soil.
Kuznetsova TA, Kam M, Khokhlova IS, Kostina NV, Dobrovolskaya TG, Umarov MM, Degen AA, Shenbrot GI, Krasnov BR. Kuznetsova TA, et al. Microb Ecol. 2013 Nov;66(4):940-9. doi: 10.1007/s00248-013-0263-7. Epub 2013 Jul 16. Microb Ecol. 2013. PMID: 23857378 - Biosorptive Potential of Pseudomonas species RY12 Toward Zinc Heavy Metal in Agriculture Soil Irrigated with Contaminated Waste Water.
Yasmin R, Zafar MS, Tahir IM, Asif R, Asghar S, Raza SK. Yasmin R, et al. Dose Response. 2022 Aug 29;20(3):15593258221117352. doi: 10.1177/15593258221117352. eCollection 2022 Jul-Sep. Dose Response. 2022. PMID: 36052270 Free PMC article. - Caulobacter species as a cause of postneurosurgical bacterial meningitis in a pediatric patient.
Bridger N, Walkty A, Crockett M, Fanella S, Nichol K, Karlowsky JA. Bridger N, et al. Can J Infect Dis Med Microbiol. 2012 Spring;23(1):e10-2. doi: 10.1155/2012/231894. Can J Infect Dis Med Microbiol. 2012. PMID: 23449318 Free PMC article. - leBIBIQBPP: a set of databases and a webtool for automatic phylogenetic analysis of prokaryotic sequences.
Flandrois JP, Perrière G, Gouy M. Flandrois JP, et al. BMC Bioinformatics. 2015 Aug 12;16(1):251. doi: 10.1186/s12859-015-0692-z. BMC Bioinformatics. 2015. PMID: 26264559 Free PMC article. - Assessment of dust sampling methods for the study of cultivable-microorganism exposure in stables.
Normand AC, Vacheyrou M, Sudre B, Heederik DJ, Piarroux R. Normand AC, et al. Appl Environ Microbiol. 2009 Dec;75(24):7617-23. doi: 10.1128/AEM.01414-09. Epub 2009 Oct 9. Appl Environ Microbiol. 2009. PMID: 19820146 Free PMC article.
References
- Ash C, Farrow J A E, Dorsch M, Stackebrandt E, Collins M D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species based on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. - PubMed
- Cilia V, Lafay B, Christen R. Sequence heterogeneities exist among the 16S ribosomal RNA sequences of the seven operons in Escherichia coli strain PK3 that can affect phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. - PubMed
- Clayton R A, Sutton G, Hinkle P S, Jr, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. - PubMed
- Dessen P, Fondrat C, Valencien C, Munier G. BISANCE: a French service for access to biomolecular sequences databases. CABIOS. 1990;6:355–356. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases