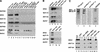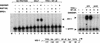Functional selectivity of recombinant mammalian SWI/SNF subunits - PubMed (original) (raw)
Functional selectivity of recombinant mammalian SWI/SNF subunits
S Kadam et al. Genes Dev. 2000.
Abstract
The SWI/SNF family of chromatin-remodeling complexes plays a key role in facilitating the binding of specific transcription factors to nucleosomal DNA in diverse organisms from yeast to man. Yet the process by which SWI/SNF and other chromatin-remodeling complexes activate specific subsets of genes is poorly understood. We show that mammalian SWI/SNF regulates transcription from chromatin-assembled genes in a factor-specific manner in vitro. The DNA-binding domains (DBDs) of several zinc finger proteins, including EKLF, interact directly with SWI/SNF to generate DNase I hypersensitivity within the chromatin-assembled beta-globin promoter. Interestingly, we find that two SWI/SNF subunits (BRG1 and BAF155) are necessary and sufficient for targeted chromatin remodeling and transcriptional activation by EKLF in vitro. Remodeling is achieved with only the BRG1-BAF155 minimal complex and the EKLF zinc finger DBD, whereas transcription requires, in addition, an activation domain. In contrast, the BRG1-BAF155 complex does not interact or function with two unrelated transcription factors, TFE3 and NF-kappaB. We conclude that specific domains of certain transcription factors differentially target SWI/SNF complexes to chromatin in a gene-selective manner and that individual SWI/SNF subunits play unique roles in transcription factor-directed nucleosome remodeling.
Figures
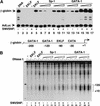
Figure 1
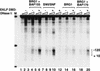
Mammalian SWI/SNF selectively functions with several zinc finger DNA-binding proteins to remodel chromatin and activate transcription in vitro. (A) In vitro transcription of chromatin-assembled β-globin plasmids by zinc finger containing transcription factors (EKLF, GATA-1, and Sp-1) in the presence of mammalian SWI/SNF chromatin-remodeling complex. β-globin plasmid templates were assembled into chromatin and incubated in the absence (lanes 1–3, 6–8, 12–14) or presence (lanes 4,5, 9–11,15–17) of SWI/SNF and, where indicated, 37 pmole of recombinant EKLF (lanes 3,5); 30 nmole, 60 nmole, and 150 nmole of an Sp1 fraction (lanes 6–11), and 35 pmole, 45 pmole, or 65 pmole of recombinant GATA-1 (lanes 12–17) per 1 μg of chromatin in a 100 μL of reaction volume. Triangles indicate increasing concentrations of Sp1 (which was inhibitory to transcription at high levels in the absence of SWI/SNF) or GATA-1. All proteins were added to assembled chromatin and incubated for 30 min at 27°C. Chromatin assembly and transcription reactions were conducted as described in Materials and Methods. Primer extension products of the β-globin promoter and the AdLuc internal control gene are indicated by arrows. (B) Analysis of the ability of different zinc finger-containing DNA-binding proteins to generate DNase I hypersensitivity within the β-globin promoter in the presence of SWI/SNF. Assembled chromatin was incubated with SWI/SNF and either EKLF, Sp1, or GATA-1 as in A, and half of the reaction was divided in two and digested with 2 U and 1 U of DNase I as described in Materials and Methods. Triangles indicate increasing amounts of DNase I. Brackets show the −120–+10 region of the β-globin promoter. A schematic diagram of the β-globin promoter is shown between panels A and B.
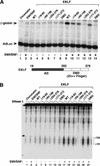
Figure 2
Distinct protein domains of EKLF are required for SWI/SNF-dependent chromatin remodeling and transcriptional activation. (A) Analysis of different EKLF mutant proteins in β-globin promoter activation in the presence or absence of SWI/SNF in vitro. Assembled chromatin templates were incubated with either wild-type or mutant EKLF proteins (37 pmole/1 μg of chromatin in a 100 μL reaction volume) and SWI/SNF, as indicated for each lane. The reactions were then split, and half was transcribed as in Figure 1A and as described in Materials and Methods. (B) The zinc finger DNA-binding domain of EKLF is sufficient to direct SWI/SNF-dependent DNase I hypersensitivity within the β-globin promoter. After assembly, chromatin was incubated with either wild-type or mutant EKLF protein in the presence or absence of SWI/SNF, as indicated above each lane. Reactions were then split and half was transcribed as shown in A and the remaining chromatin was divided into two tubes with 150 ng chromatin per tube and digested with 1 U and 2 U of DNase I. Triangles indicate increasing amounts of DNase I. Brackets show the −120–+10 region of the promoter. (M) Digested β-globin plasmid as a size marker. Bands represent digests of _Nco_I (4.7 Kb), _Nco_I/_Bsa_AI (841 bp) and _Nco_I/_Eco_RI (301 bp). A schematic diagram of the domain structure of EKLF is shown between panels A and B.
Figure 3
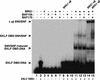
SWI/SNF subunits interact specifically with the zinc finger DNA-binding domains (DBDs) of EKLF, GATA-1, and Sp1. (A) Mammalian SWI/SNF interacts with zinc finger DBDs. GST pull-down assays were performed with 3 μg SWI/SNF and 1 μg GST-fused wild-type or mutant EKLF, GATA-1, TFE3, NF-κB (p50). SWI/SNF subunits were detected by Western blot analyses using the antisera indicated on the left as described in Materials and Methods. (B) Acidic and proline-rich activation domains do not interact with mammalian SWI/SNF. GST pull-down assays were performed using 1.5 μg of SWI/SNF and 500 ng each of GST-fused wild-type or mutant EKLF and VP16 proteins. Histidine pull-down assays were performed using 500 ng each of wild-type EKLF, EKLF DBD, wild-type GAL4–VP16, and GAL4 DBD as described in Materials and Methods. Proteins that were pulled down with the beads were analyzed on a 10% SDS-PAGE gel and immunoblotted with antibodies against SWI/SNF subunits, BRG1, BAF170, BAF155, and BAF57. (C) Specific SWI/SNF subunits interact with the EKLF zinc finger DBD. (Top panel) Flag-tagged hSWI/SNF was analyzed by SDS-PAGE, stained with silver (lane 4), blotted onto a PVDF membrane, and processed for far-Western analysis using three different probes: GST–EKLF, followed by anti-EKLF antibody (lane 2), 32P-labeled GST–EKLF (lane 1), or GST–EKLF (AD) followed by anti-EKLF antibody (lane 3) as described in Materials and Methods. The positions of SWI/SNF subunits, BRG1, BAF170, and BAF155 are indicated. (Bottom panel) Wild-type EKLF and the EKLF DBD interact with recombinant SWI/SNF subunits. GST pull-down assays were carried out using 200 ng of purified recombinant F-BRG1, F-BAF170, or F-BAF155 with 200 ng of GST-fused wild-type or mutant EKLF and Sp1 proteins. Equal amounts of supernatants (S) and beads (B) were analyzed on a 10% SDS-PAGE gel and immunoblotted with antibodies against SWI/SNF subunits, BRG1, BAF170, and BAF155.
Figure 4
A minimal recombinant SWI/SNF complex is sufficient for EKLF-dependent transcriptional activation of chromatin-assembled β-globin genes. Recombinant BRG1 and BAF155 or BAF170 cooperate with EKLF to activate chromatin-assembled β-globin genes in vitro. After nucleosome assembly, 100 ng of chromatin was incubated with: 3.7 pmole of EKLF as indicated, 20 ng of F-BRG1 (lanes 4,5,10–16,18–19,21–22), 140 ng of F-BAF170 (lanes 8–9,14–15,17–19), 100 ng of F-BAF155 (lanes 10–11), and 140 ng of F-BAF155 (lanes 12–15,20–22), followed by primer extension analysis of transcripts. As a positive control, an aliquot of 58 ng purified SWI/SNF was incubated with 3.7 pmole of EKLF (lane 3).
Figure 5
Recombinant SWI/SNF subunits do not support transcription from chromatin-assembled HIV-1 promoters by TFE3 and NF-κB. Factor-dependent transcriptional activation by native and recombinant SWI/SNF on a chromatin-assembled HIV-1 promoter. (Left ) an aliquot of 100 ng pHIV-1–Luc was assembled into chromatin in the absence of transcription factors (lanes 1–5), or with the following recombinant proteins: 4 pmole EKLF (lanes 6–8) or 5 pmole TFE3 plus one pmole NF-κB subunits (p50:p65; lanes 9–13). Where indicated, 20 ng F-BRG1, 140 ng F-BAF155, and 58 ng native SWI/SNF were added after nucleosome assembly. (Right) A mixture of 5 pmole TFE3 and 1 pmole NF-κB (p50:p65) was incubated with 100 ng HIV-1 chromatin either before (lanes 16–17) or after (lanes 18–19) nucleosome assembly. Transcription reactions contained (lanes 15,17,19) or lacked (lanes 14,16,18) 58 ng native SWI/SNF. The α-globin gene was included as a control for transcription and RNA recovery. A schematic diagram of the HIV-1 promoter is shown below.
Figure 6
The EKLF zinc finger DNA-binding domain (DBD) is sufficient to target chromatin remodeling by the BRG1–BAF155 minimal complex. Analysis of recombinant SWI/SNF subunits to generate DNase I hypersensitive sites within the chromatin-assembled β-globin promoter in the presence of the EKLF DBD. After nucleosome assembly, 300 ng of chromatin was incubated with the following proteins as indicated: recombinant SWI/SNF subunits (60 ng F-BRG1, 400 ng F-BAF155, 400 ng F-BAF170); native SWI/SNF (180 ng); and EKLF DBD (11 pmole). 100 ng of the reaction was transcribed as a control (data not shown) and the remaining chromatin was divided in half and digested with 2 U and 1 U of DNase I as described in Materials and Methods. Triangles indicate increasing amounts of DNase I. (M) Digested β-globin plasmid used as a size marker. Bands represent digests of _Nco_I (4.7 kb), _Nco_I/_Bsa_AI (841 bp) and _Nco_I/_Eco_RI (301 bp).
Figure 7
Interaction of recombinant SWI/SNF subunits with the EKLF DNA-binding domain (DBD) and DNA. A 60-bp region of the β-globin promoter containing the EKLF binding site was incubated with the following proteins: 10 ng of EKLF DBD (lanes 2,8–15); 20 ng F-BRG1 (lanes 3,6–7,11–15); 140 ng of F-BAF155 (lanes 4,6,9,12,14); 140 ng of F-BAF170 (lanes 5,7,10,13,15) and analyzed by EMSA. One microliter of undiluted antibodies was added as indicated.
Figure 8
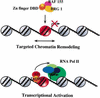
Model for targeted chromatin remodeling and transcriptional activation, highlighting the interaction between the EKLF zinc finger DNA-binding domain and mammalian SWI/SNF subunits.
Similar articles
- A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro.
Armstrong JA, Bieker JJ, Emerson BM. Armstrong JA, et al. Cell. 1998 Oct 2;95(1):93-104. doi: 10.1016/s0092-8674(00)81785-7. Cell. 1998. PMID: 9778250 - Distinct domains of erythroid Krüppel-like factor modulate chromatin remodeling and transactivation at the endogenous beta-globin gene promoter.
Brown RC, Pattison S, van Ree J, Coghill E, Perkins A, Jane SM, Cunningham JM. Brown RC, et al. Mol Cell Biol. 2002 Jan;22(1):161-70. doi: 10.1128/MCB.22.1.161-170.2002. Mol Cell Biol. 2002. PMID: 11739731 Free PMC article. - Novel Interactions between the Human T-Cell Leukemia Virus Type 1 Antisense Protein HBZ and the SWI/SNF Chromatin Remodeling Family: Implications for Viral Life Cycle.
Alasiri A, Abboud Guerr J, Hall WW, Sheehy N. Alasiri A, et al. J Virol. 2019 Jul 30;93(16):e00412-19. doi: 10.1128/JVI.00412-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31142665 Free PMC article. - BAFfling pathologies: Alterations of BAF complexes in cancer.
Arnaud O, Le Loarer F, Tirode F. Arnaud O, et al. Cancer Lett. 2018 Apr 10;419:266-279. doi: 10.1016/j.canlet.2018.01.046. Epub 2018 Jan 31. Cancer Lett. 2018. PMID: 29374542 Review. - Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence.
Montecino M, Stein JL, Stein GS, Lian JB, van Wijnen AJ, Cruzat F, Gutiérrez S, Olate J, Marcellini S, Gutiérrez JL. Montecino M, et al. Biochem Cell Biol. 2007 Aug;85(4):419-25. doi: 10.1139/O07-070. Biochem Cell Biol. 2007. PMID: 17713577 Review.
Cited by
- EKLF/KLF1, a tissue-restricted integrator of transcriptional control, chromatin remodeling, and lineage determination.
Yien YY, Bieker JJ. Yien YY, et al. Mol Cell Biol. 2013 Jan;33(1):4-13. doi: 10.1128/MCB.01058-12. Epub 2012 Oct 22. Mol Cell Biol. 2013. PMID: 23090966 Free PMC article. Review. - Coordination of cell signaling, chromatin remodeling, histone modifications, and regulator recruitment in human matrix metalloproteinase 9 gene transcription.
Ma Z, Shah RC, Chang MJ, Benveniste EN. Ma Z, et al. Mol Cell Biol. 2004 Jun;24(12):5496-509. doi: 10.1128/MCB.24.12.5496-5509.2004. Mol Cell Biol. 2004. PMID: 15169910 Free PMC article. - Vulnerabilities of mutant SWI/SNF complexes in cancer.
Helming KC, Wang X, Roberts CWM. Helming KC, et al. Cancer Cell. 2014 Sep 8;26(3):309-317. doi: 10.1016/j.ccr.2014.07.018. Cancer Cell. 2014. PMID: 25203320 Free PMC article. Review. - GBAF, a small BAF sub-complex with big implications: a systematic review.
Innis SM, Cabot B. Innis SM, et al. Epigenetics Chromatin. 2020 Nov 3;13(1):48. doi: 10.1186/s13072-020-00370-8. Epigenetics Chromatin. 2020. PMID: 33143733 Free PMC article. - Endometriosis-associated ovarian carcinomas: insights into pathogenesis, diagnostics, and therapeutic targets-a narrative review.
Samartzis EP, Labidi-Galy SI, Moschetta M, Uccello M, Kalaitzopoulos DR, Perez-Fidalgo JA, Boussios S. Samartzis EP, et al. Ann Transl Med. 2020 Dec;8(24):1712. doi: 10.21037/atm-20-3022a. Ann Transl Med. 2020. PMID: 33490224 Free PMC article. Review.
References
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous