Replication past O(6)-methylguanine by yeast and human DNA polymerase eta - PubMed (original) (raw)
Replication past O(6)-methylguanine by yeast and human DNA polymerase eta
L Haracska et al. Mol Cell Biol. 2000 Nov.
Abstract
O(6)-Methylguanine (m6G) is formed by the action of alkylating agents such as N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) on DNA. m6G is a highly mutagenic and carcinogenic lesion, and it presents a block to synthesis by DNA polymerases. Here, we provide genetic and biochemical evidence for the involvement of yeast and human DNA polymerase eta (Poleta) in the replicative bypass of m6G lesions in DNA. The formation of MNNG-induced mutations is almost abolished in the rad30Delta pol32Delta double mutant of yeast, which lacks the RAD30 gene that encodes Poleta and the Pol32 subunit of DNA polymerase delta (Poldelta). Although Poldelta can function in the mutagenic bypass of m6G lesions, our biochemical studies indicate that Poleta is much more efficient in replicating through m6G than Poldelta. Both Poleta and Poldelta insert a C or a T residue opposite from m6G; Poleta, however, is more accurate, as it inserts a C about twice as frequently as Poldelta. Alkylating agents are used in the treatment of malignant tumors, including lymphomas, brain tumors, melanomas, and gastrointestinal carcinomas, and the clinical effectiveness of these agents derives at least in part from their ability to form m6G in DNA. Inactivation of Poleta could afford a useful strategy for enhancing the effectiveness of these agents in cancer chemotherapy.
Figures
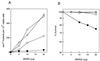
FIG. 1
MNNG-induced mutations at the CAN1 locus in _rad30_Δ and _pol32_Δ yeast strains. _can1_r mutation frequency (A) and viability (B) in MNNG-treated yeast strains are shown. Cells grown overnight in YPD medium were treated with MNNG at the concentrations indicated for a 20-min period. Appropriate dilutions were spread onto synthetic complete medium lacking arginine and containing canavanine for the determination of _CAN1_s to _can1_r mutation frequencies and onto YPD plates for viability determinations. Each curve represents the average of two or more experiments. ▵, EMY74.7 (wild type) (RAD30 POL32); ○, YPO-69 (_pol32_Δ); □, YR30.2 (_rad30_Δ); ■, YR30.97 (_rad30Δ pol32_Δ).
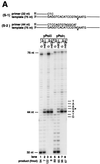
FIG. 2
Translesion DNA synthesis by yeast Polη and yeast Polδ on templates containing m6G. (A) Running-start and standing-start DNA synthesis past m6G by yeast Polη and yeast Polδ. Sequences adjacent to the primer:template junction are shown for 75-nt template and 32-nt (S-1 substrate, lanes 1, 2, 5, and 6) or 44-nt (S-2 substrate, lanes 3, 4, 7, and 8) primer. The primers were 32P-labeled at their 5′ end. The position of undamaged G (lanes 1, 3, 5, and 7) or m6G (lanes 2, 4, 6, and 8) on the template is indicated by G*. Yeast Polδ (10 nM) (lanes 1 to 4) or yeast Polη (2.5 nM) (lanes 5 to 8) was incubated with the DNA substrate (20 nM) for 5 min at 30°C. Reaction products were resolved on a 10% denaturing polyacrylamide gel and were visualized by autoradiography. The amount of synthesis past the undamaged G or m6G is indicated. (B) Nucleotides incorporated opposite m6G by yeast Polη and yeast Polδ. Standing-start reactions were carried out on a G (lanes 5 and 7)- or m6G (lanes 6 and 8)-containing 18-nt template primed with a 5′ 32P-labeled 12-nt oligomer. The position corresponding to the G or m6G residue in the template is indicated by G*. Reactions were carried out as described for panel A above, except that the following DNA polymerase concentrations were used: yeast Polδ, 40 nM (lanes 5 and 6); and yeast Polη, 10 nM (lanes 7 and 8). Reaction mixtures were resolved on 20% denaturing polyacrylamide gel, and electrophoretic mobilities of the 18-nt reaction products (lanes 5 to 8) were compared with those of 18-nt synthetic oligomers containing a G (lane 1), a T (lane 2), an A (lane 3), or a C (lane 4) residue at position 13.
FIG. 2
Translesion DNA synthesis by yeast Polη and yeast Polδ on templates containing m6G. (A) Running-start and standing-start DNA synthesis past m6G by yeast Polη and yeast Polδ. Sequences adjacent to the primer:template junction are shown for 75-nt template and 32-nt (S-1 substrate, lanes 1, 2, 5, and 6) or 44-nt (S-2 substrate, lanes 3, 4, 7, and 8) primer. The primers were 32P-labeled at their 5′ end. The position of undamaged G (lanes 1, 3, 5, and 7) or m6G (lanes 2, 4, 6, and 8) on the template is indicated by G*. Yeast Polδ (10 nM) (lanes 1 to 4) or yeast Polη (2.5 nM) (lanes 5 to 8) was incubated with the DNA substrate (20 nM) for 5 min at 30°C. Reaction products were resolved on a 10% denaturing polyacrylamide gel and were visualized by autoradiography. The amount of synthesis past the undamaged G or m6G is indicated. (B) Nucleotides incorporated opposite m6G by yeast Polη and yeast Polδ. Standing-start reactions were carried out on a G (lanes 5 and 7)- or m6G (lanes 6 and 8)-containing 18-nt template primed with a 5′ 32P-labeled 12-nt oligomer. The position corresponding to the G or m6G residue in the template is indicated by G*. Reactions were carried out as described for panel A above, except that the following DNA polymerase concentrations were used: yeast Polδ, 40 nM (lanes 5 and 6); and yeast Polη, 10 nM (lanes 7 and 8). Reaction mixtures were resolved on 20% denaturing polyacrylamide gel, and electrophoretic mobilities of the 18-nt reaction products (lanes 5 to 8) were compared with those of 18-nt synthetic oligomers containing a G (lane 1), a T (lane 2), an A (lane 3), or a C (lane 4) residue at position 13.
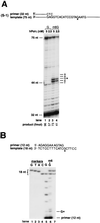
FIG. 3
Translesion DNA synthesis activity of hPolη on template containing m6G. (A) Running-start DNA synthesis past m6G by hPolη. The position of undamaged G or the corresponding m6G is indicated by G*. Five nanomolar (lanes 1 and 3) or 2.5 nM (lanes 2 and 4) hPolη was incubated with DNA substrate (20 nM) for 5 min at 30°C under standard reaction conditions. Undamaged DNA template, lanes 1 and 2; m6G template, lanes 3 and 4. (B) Deoxynucleotide incorporation opposite m6G by hPolη. The position of the m6G or the undamaged G residue in template DNA is indicated by G*. hPolη (10 nM) was incubated with 20 nM undamaged (lane 5) or m6G-containing (lane 6) DNA substrate under standard reaction conditions. Electrophoretic mobilities of 18-nt reaction products (lanes 5 and 6) were compared with those of 18-nt synthetic oligomer markers containing a G, T, A, or C residue at position 13 (lanes 1 to 4, respectively). Two of these 18-nt marker oligomers, containing a C (lower band) or T (upper band) at position 13, were mixed and run in lane 7.
Similar articles
- Role of DNA polymerase eta in the bypass of abasic sites in yeast cells.
Zhao B, Xie Z, Shen H, Wang Z. Zhao B, et al. Nucleic Acids Res. 2004 Jul 29;32(13):3984-94. doi: 10.1093/nar/gkh710. Print 2004. Nucleic Acids Res. 2004. PMID: 15284331 Free PMC article. - Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine.
Haracska L, Prakash S, Prakash L. Haracska L, et al. Mol Cell Biol. 2003 Feb;23(4):1453-9. doi: 10.1128/MCB.23.4.1453-1459.2003. Mol Cell Biol. 2003. PMID: 12556503 Free PMC article. - Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta.
Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Haracska L, et al. Nat Genet. 2000 Aug;25(4):458-61. doi: 10.1038/78169. Nat Genet. 2000. PMID: 10932195 - POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies.
Nicolas E, Golemis EA, Arora S. Nicolas E, et al. Gene. 2016 Sep 15;590(1):128-41. doi: 10.1016/j.gene.2016.06.031. Epub 2016 Jun 16. Gene. 2016. PMID: 27320729 Free PMC article. Review.
Cited by
- Trans-lesion synthesis and mismatch repair pathway crosstalk defines chemoresistance and hypermutation mechanisms in glioblastoma.
Cheng X, An J, Lou J, Gu Q, Ding W, Droby GN, Wang Y, Wang C, Gao Y, Anand JR, Shelton A, Satterlee AB, Mann B, Hsiao YC, Liu CW, Lu K, Hingtgen S, Wang J, Liu Z, Miller CR, Wu D, Vaziri C, Yang Y. Cheng X, et al. Nat Commun. 2024 Mar 4;15(1):1957. doi: 10.1038/s41467-024-45979-5. Nat Commun. 2024. PMID: 38438348 Free PMC article. - For the Better or for the Worse? The Effect of Manganese on the Activity of Eukaryotic DNA Polymerases.
Balint E, Unk I. Balint E, et al. Int J Mol Sci. 2023 Dec 27;25(1):363. doi: 10.3390/ijms25010363. Int J Mol Sci. 2023. PMID: 38203535 Free PMC article. Review. - Trans-Lesion Synthesis and Mismatch Repair Pathway Crosstalk Defines Chemoresistance and Hypermutation Mechanisms in Glioblastoma.
Cheng X, An J, Lou J, Gu Q, Ding W, Droby G, Wang Y, Wang C, Gao Y, Shelton A, Satterlee AB, Mann BE, Hsiao YC, Liu CW, Liu K, Hingtgen S, Wang J, Liu Z, Miller R, Wu D, Vaziri C, Yang Y. Cheng X, et al. bioRxiv [Preprint]. 2023 Oct 19:2023.10.16.562506. doi: 10.1101/2023.10.16.562506. bioRxiv. 2023. PMID: 37905107 Free PMC article. Updated. Preprint. - DNA Alkylation Damage by Nitrosamines and Relevant DNA Repair Pathways.
Fahrer J, Christmann M. Fahrer J, et al. Int J Mol Sci. 2023 Feb 28;24(5):4684. doi: 10.3390/ijms24054684. Int J Mol Sci. 2023. PMID: 36902118 Free PMC article. Review.
References
- Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. - PubMed
- Ciarrocchi G, Pedrini A M. Determination of pyrimidine dimer unwinding angle by measurement of DNA electrophoretic mobility. J Mol Biol. 1982;155:177–183. - PubMed
- Creighton S, Bloom L B, Goodman M F. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. - PubMed
- Day R S, III, Ziolkowski C H J, Scudiero D A, Meyer S A, Mattern M R. Human tumor cell strains defective in the repair of alkylation damage. Carcinogenesis. 1980;1:21–32. - PubMed
- de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous