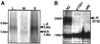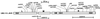Expression of multiple UNC-13 proteins in the Caenorhabditis elegans nervous system - PubMed (original) (raw)
Expression of multiple UNC-13 proteins in the Caenorhabditis elegans nervous system
R E Kohn et al. Mol Biol Cell. 2000 Oct.
Free PMC article
Abstract
The Caenorhabditis elegans UNC-13 protein and its mammalian homologues are important for normal neurotransmitter release. We have identified a set of transcripts from the unc-13 locus in C. elegans resulting from alternative splicing and apparent alternative promoters. These transcripts encode proteins that are identical in their C-terminal regions but that vary in their N-terminal regions. The most abundant protein form is localized to most or all synapses. We have analyzed the sequence alterations, immunostaining patterns, and behavioral phenotypes of 31 independent unc-13 alleles. Many of these mutations are transcript-specific; their phenotypes suggest that the different UNC-13 forms have different cellular functions. We have also isolated a deletion allele that is predicted to disrupt all UNC-13 protein products; animals homozygous for this null allele are able to complete embryogenesis and hatch, but they die as paralyzed first-stage larvae. Transgenic expression of the entire gene rescues the behavior of mutants fully; transgenic overexpression of one of the transcripts can partially compensate for the genetic loss of another. This finding suggests some degree of functional overlap of the different protein products.
Figures
Figure 1
Genomic region around unc-13, with predicted unc-13 transcripts and protein products. (A) Genomic sequence indicating the locations of the major_unc-13_ transcript and the extents of cosmids C44E1 and ZK524. (B) Genomic sequence with introns and exons indicating the production of three predicted unc-13 transcripts. Region_L_ (left) corresponds to the less conserved 5′ end of_unc-13_, and region R (right) corresponds to the more highly conserved 3′ end of unc-13. Region_M_ (middle) corresponds to an exon that was not originally identified in the published sequence of_unc-13_. Asterisks mark positions of alternative splicing. (C) Proteins expressed from the predicted transcripts are shown. cDNA library screens and RACE products indicate the existence of_unc-13_ products. The cDNAs yk50c3 (GenBank accession number D67349), yk16b7 (GenBank accession numbers D26913 and D35032), and yk8h12 (GenBank accession number D37721) were isolated by Yuji Kohara from a dT-primed C. elegans library. The remaining cDNAs (RM#418p, RM#384p, RM#419p, RM#385p, and RM#386p) were isolated in our laboratory from a random primed C. elegans library. 5′ RACE and cRACE (Maruyama et al., 1995) were used to produce RACE products.
Figure 2
UNC-13 has several mammalian homologues. The UNC-13L-R structure is shown. This form is similar to the published form of UNC-13 (Maruyama and Brenner, 1991). Additional N-terminal regions, including a third C2 domain, were identified in cDNA library screens (GenBank accession number M62830). C1 and C2 represent PKC homology domains. Munc13-1, Munc13-2, and Munc13-3 (GenBank accession numbers U24070, U24071, and U75261, respectively) are rat homologues of_C. elegans_ UNC-13 (Brose et al., 1995). Hmunc13 (GenBank accession number AF020202) is a human homologue of UNC-13 (Song et al., 1999). The UNC-13L region is 30% identical to the N-terminal region of Munc13-1.
Figure 3
Northern and Western transfers show products of the unc-13 gene. (A) Northern transfers indicate the presence of unc-13L-R and unc-13M-R transcripts. L, M, and R indicate lanes incubated with32P-labeled probes that hybridize specifically to the_unc-13L_, unc-13M, and_unc-13R_ regions, respectively. (B) Western transfers indicate the presence of several high-molecular-mass protein products from the unc-13 gene. Proteins from N2,unc-13(e1091), and unc-13(s69) bound to nitrocellulose membrane were labeled with anti-UNC-13L antibody. The band present at ∼130 kDa is nonspecific.
Figure 4
Sequenced unc-13 alleles. The positions of mutations are indicated on a diagram of an UNC-13L-M-R protein. Alleles with names shown in bold encode in-frame (i.e., missense) mutations. The unc-13L region (L) includes exons 1–13; unc-13M (M) consists of exon 14; and_unc-13R_ (R) includes exons 15–31.
Figure 5
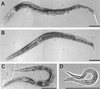
Appearance of unc-13 mutants. (A) N2 (wild-type) young adult. (B) Young adult homozygous for the severe_L_-region mutation e1091. (C) Young adult homozygous for the severe R_-region mutation_s69. (D) Recently hatched larva homozygous for the lethal mutation md2415. These animals do not grow; they barely move, and they die within a few days. Bars, 100 μm.
Figure 6
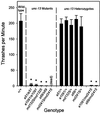
unc-13 behavioral phenotypes are recessive, and L and R mutants fail to complement one another. Thrashing values are body bends per minute measured during a 2-min interval; values are means of 10–20 nematodes ± SDs. The wild-type data are representative of tests on a single day. Probabilities were calculated with the use of Student's t test for each strain versus a matched set of wild-type nematodes. *Significantly different from wild-type (p < 0.001). The homozygous unc-13 alleles (e51, e1091, md1325, and_s69_) are not significantly different from each other or from e1091/md2415 or s69/md2415.
Figure 7
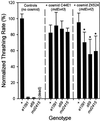
Behavioral phenotypes of transgenic strains. Transgenic arrays including the cosmid ZK524 (mdEx42) or the cosmid C44E1 (mdEx43) were generated by injection of wild-type nematodes. The arrays were crossed into the_unc-13_ mutants e1091, s69, and md2415. All strains were assayed for thrashing in liquid. Values given are normalized to N2 values measured on the same day. *Significantly different from N2 plus ZK524 (p < 0.01).
Figure 8
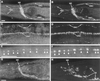
Anti-UNC-13L antibody stains a form of UNC-13 protein that is synaptic but is unlikely to be on synaptic vesicles. C. elegans were double stained with anti-UNC-13L (left) and anti-UNC-17/VAChT (right). (A) Anti-UNC-13L in an adult wild-type head stains the major synaptic regions, including the nerve ring (NR), ventral nerve cord (VNC), dorsal nerve cord (DNC), and sublateral nerve cords (SNC). (B) Anti-UNC-17/VAChT stains a subset of the region stained by anti-UNC-13L. (C and D) A slightly higher magnification shows punctate staining with both antibodies in the dorsal nerve cord and sublateral nerve cords in the body. (E and F) A higher magnification shows the colocalization of anti-UNC-13L (E) and anti-UNC-17/VAChT (F) in synaptic regions (arrowheads) in one of the sublateral nerve cords in the head. (G and H) In an adult_unc-104_ mutant, staining of anti-UNC-13L is somewhat less intense but remains synaptic, whereas anti-UNC-17/VAChT is now localized in neuronal cell bodies (CB). Anterior is left; bars, 10 μ m.
Similar articles
- Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system.
Maduro M, Pilgrim D. Maduro M, et al. Genetics. 1995 Nov;141(3):977-88. doi: 10.1093/genetics/141.3.977. Genetics. 1995. PMID: 8582641 Free PMC article. - Alternative splicing leads to two cholinergic proteins in Caenorhabditis elegans.
Alfonso A, Grundahl K, McManus JR, Asbury JM, Rand JB. Alfonso A, et al. J Mol Biol. 1994 Aug 26;241(4):627-30. doi: 10.1006/jmbi.1994.1538. J Mol Biol. 1994. PMID: 8057385 - The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin.
Saifee O, Wei L, Nonet ML. Saifee O, et al. Mol Biol Cell. 1998 Jun;9(6):1235-52. doi: 10.1091/mbc.9.6.1235. Mol Biol Cell. 1998. PMID: 9614171 Free PMC article. - Mammalian Unc-13 homologues as possible regulators of neurotransmitter release.
Betz A, Telemenakis I, Hofmann K, Brose N. Betz A, et al. Biochem Soc Trans. 1996 Aug;24(3):661-6. doi: 10.1042/bst0240661. Biochem Soc Trans. 1996. PMID: 8878822 Review. No abstract available. - UNC-52/perlecan isoform diversity and function in Caenorhabditis elegans.
Rogalski TM, Mullen GP, Bush JA, Gilchrist EJ, Moerman DG. Rogalski TM, et al. Biochem Soc Trans. 2001 May;29(Pt 2):171-6. doi: 10.1042/0300-5127:0290171. Biochem Soc Trans. 2001. PMID: 11356148 Review.
Cited by
- Low-dose naltrexone extends healthspan and lifespan in C. elegans via SKN-1 activation.
Li W, McIntyre RL, Schomakers BV, Kamble R, Luesink AHG, van Weeghel M, Houtkooper RH, Gao AW, Janssens GE. Li W, et al. iScience. 2024 May 8;27(6):109949. doi: 10.1016/j.isci.2024.109949. eCollection 2024 Jun 21. iScience. 2024. PMID: 38799567 Free PMC article. - Expression and function of Caenorhabditis elegans UNCP-18, a paralog of the SM protein UNC-18.
Boeglin M, Leyva-Díaz E, Hobert O. Boeglin M, et al. Genetics. 2023 Dec 6;225(4):iyad180. doi: 10.1093/genetics/iyad180. Genetics. 2023. PMID: 37793339 Free PMC article. - Functional Roles of UNC-13/Munc13 and UNC-18/Munc18 in Neurotransmission.
Meunier FA, Hu Z. Meunier FA, et al. Adv Neurobiol. 2023;33:203-231. doi: 10.1007/978-3-031-34229-5_8. Adv Neurobiol. 2023. PMID: 37615868 Review. - Muscle-directed mechanosensory feedback activates egg-laying circuit activity and behavior in Caenorhabditis elegans.
Medrano E, Collins KM. Medrano E, et al. Curr Biol. 2023 Jun 5;33(11):2330-2339.e8. doi: 10.1016/j.cub.2023.05.008. Epub 2023 May 25. Curr Biol. 2023. PMID: 37236183 Free PMC article. - C. elegans Clarinet/CLA-1 recruits RIMB-1/RIM-binding protein and UNC-13 to orchestrate presynaptic neurotransmitter release.
Krout M, Oh KH, Xiong A, Frankel EB, Kurshan PT, Kim H, Richmond JE. Krout M, et al. Proc Natl Acad Sci U S A. 2023 May 23;120(21):e2220856120. doi: 10.1073/pnas.2220856120. Epub 2023 May 15. Proc Natl Acad Sci U S A. 2023. PMID: 37186867 Free PMC article.
References
- Alfonso A, Grundahl K, Duerr JS, Han H-P, Rand JB. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science. 1993;261:617–619. - PubMed
- Barstead RJ. Reverse genetics. In: Hope IA, editor. C. elegans: A Practical Approach. Oxford, UK: Oxford University Press; 1999. pp. 97–118.
- Betz A, Ashery U, Rickmann M, Augustin I, Neher E, Südhof TC, Rettig J, Brose N. Munc13-1 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron. 1998;21:123–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases