Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays - PubMed (original) (raw)
Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays
S Peterson et al. J Bacteriol. 2000 Nov.
Abstract
Competence for genetic transformation in Streptococcus pneumoniae is coordinated by the competence-stimulating peptide (CSP), which induces a sudden and transient appearance of competence during exponential growth in vitro. Models of this quorum-sensing mechanism have proposed sequential expression of several regulatory genes followed by induction of target genes encoding DNA-processing-pathway proteins. Although many genes required for transformation are known to be expressed only in response to CSP, the relative timing of their expression has not been established. Overlapping expression patterns for the genes cinA and comD (G. Alloing, B. Martin, C. Granadel, and J. P. Claverys, Mol. Microbiol. 29:75-83, 1998) suggest that at least two distinct regulatory mechanisms may underlie the competence cycle. DNA microarrays were used to estimate mRNA levels for all known competence operons during induction of competence by CSP. The known competence regulatory operons, comAB, comCDE, and comX, exhibited a low or zero initial (uninduced) signal, strongly increased expression during the period between 5 and 12 min after CSP addition, and a decrease nearly to original values by 15 min after initiation of exposure to CSP. The remaining competence genes displayed a similar expression pattern, but with an additional delay of approximately 5 min. In a mutant defective in ComX, which may act as an alternate sigma factor to allow expression of the target competence genes, the same regulatory genes were induced, but the other competence genes were not. Finally, examination of the expression of 60 candidate sites not previously associated with competence identified eight additional loci that could be induced by CSP.
Figures
FIG. 1
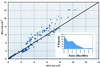
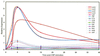
Correlation between relative message abundance measurements by hybridization with two independently labeled cDNA preparations. A scatter plot is shown of the pairs of Cy3 and Cy5 fluorescence values obtained by hybridization to a 40-gene microarray containing elements spotted at least 16 times each. cDNA was prepared from RNA (2 μg) isolated 10 min after exposure to CSP and was independently labeled with either Cy3 or Cy5 and subsequently combined for hybridization. The signal strength for Cy3 (550 nm) versus that for Cy5 (650 nm) is shown for each of 283 individual spots after normalization to yield 16S rRNA ratios of 1.0. The inset shows the frequency distribution of experimental error as the ratio of the larger signal divided by the smaller signal. RFU, relative fluorescence units.
FIG. 2
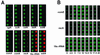
Hybridization images from representative sections of the 40-gene microarray. (A) Composite images of six replicate spots for the indicated competence genes and the control spots for ssbA and 16S rRNA. The left part of each box (Cy3; green) represents message abundance in cells 10 min post-CSP stimulation; the right side of each box (Cy5; red) represents message abundance in cells not exposed to CSP. (B) Differential temporal expression patterns of early and late competence-regulated genes. Quadruplicate spots for individual early (comE), late (recA), and control (16S rRNA) genes are shown after hybridization with cDNA prepared from RNA extracted at the indicated times (in minutes) after CSP treatment.
FIG. 3
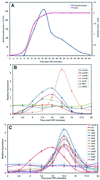
Expression patterns of competence regulons following CSP treatment of strain CP1250. (A) Competence induction monitored as capacity for DNA uptake and recombination (Novr transformants) and expression from a comX::lacZ reporter fusion in a parallel culture of CPM16. (B) mRNA levels for class 1 and class 2 genes assayed by hybridization to the 40-gene microarray. The control genes not affected by CSP treatment are ftsH and ssbA. recA is a representative of the class 3 pattern. (C) mRNA levels for class 3 genes. comA is a representative of class 2 genes. The control genes shown are ftsH, ssbA, and rpsF. Cy3-labeled cDNA was prepared from RNA isolated at the indicated times after induction of strain CP1250 with CSP. The hybridization reference, prepared from an equal-mass mixture of six RNA preparations (0 to 15 min), was labeled with Cy5.
FIG. 4
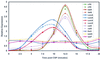
Reproducibility of expression patterns. Five CSP-inducible genes were each represented as two different PCR products on the 40-gene microarray as shown in Table 1; corresponding pairs of curves (solid and dashed) have the same color. The control genes shown (ssbA, ftsH, and rpsF) were represented twice on the microarray but with a single PCR product each. The cDNA probes were as for Fig. 3.
FIG. 5
Effect of ComX deficiency on expression patterns of CSP-inducible genes. Strain CPM4 was treated with CSP at an optical density at 550 nm of 0.04. RNA extracts were prepared from samples taken 1 min before and 2.5, 5, 15, and 40 min after CSP addition. Cy3-labeled cDNA prepared from these RNA samples was analyzed by hybridization to the 40-gene microarray using a reference Cy5 cDNA mixture prepared from RNA isolated from wild-type cells at 0, 5, 10, 15, 20, 30, and 45 min after treatment with CSP.
FIG. 6
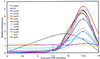
Expression patterns of candidate combox genes following CSP treatment. Hybridizations to a 68-gene microarray were performed with cDNA prepared from the same RNA extracts as in Fig. 3 and with the same hybridization reference as for Fig. 3. The profiles include controls, two uninducible candidates, and eight candidates with expression changes of threefold or more.
FIG. 7
Combox consensus sites. (A) Alignment of consensus sequences of CSP-inducible sites previously identified. TYG is induced by CSP but is not associated with a known protein product or function (5). (B) Alignment of candidate combox sites identified by pattern searches and validated as CSP inducible by expression measurements (Fig. 6). Regions (100 bp) spanning each consensus sequence were extracted and aligned with no gaps (consensus −10 and −25 elements are in boldface; mismatches to the consensus sequence are lowercase). When present, the first ATG codons of downstream ORFs are underlined.
Similar articles
- Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays.
Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. Peterson SN, et al. Mol Microbiol. 2004 Feb;51(4):1051-70. doi: 10.1046/j.1365-2958.2003.03907.x. Mol Microbiol. 2004. PMID: 14763980 - Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation.
Lee MS, Morrison DA. Lee MS, et al. J Bacteriol. 1999 Aug;181(16):5004-16. doi: 10.1128/JB.181.16.5004-5016.1999. J Bacteriol. 1999. PMID: 10438773 Free PMC article. - Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells.
Dagkessamanskaia A, Moscoso M, Hénard V, Guiral S, Overweg K, Reuter M, Martin B, Wells J, Claverys JP. Dagkessamanskaia A, et al. Mol Microbiol. 2004 Feb;51(4):1071-86. doi: 10.1111/j.1365-2958.2003.03892.x. Mol Microbiol. 2004. PMID: 14763981 - Transformation in Streptococcus pneumoniae: mosaic genes and the regulation of competence.
Hakenbeck R. Hakenbeck R. Res Microbiol. 2000 Jul-Aug;151(6):453-6. doi: 10.1016/s0923-2508(00)00170-4. Res Microbiol. 2000. PMID: 10961458 Review. - Regulation of competence for genetic transformation in Streptococcus pneumoniae: a link between quorum sensing and DNA processing genes.
Morrison DA, Lee MS. Morrison DA, et al. Res Microbiol. 2000 Jul-Aug;151(6):445-51. doi: 10.1016/s0923-2508(00)00171-6. Res Microbiol. 2000. PMID: 10961457 Review.
Cited by
- LytF, a novel competence-regulated murein hydrolase in the genus Streptococcus.
Berg KH, Ohnstad HS, Håvarstein LS. Berg KH, et al. J Bacteriol. 2012 Feb;194(3):627-35. doi: 10.1128/JB.06273-11. Epub 2011 Nov 28. J Bacteriol. 2012. PMID: 22123253 Free PMC article. - Comparison of Surface Proteomes of Adherence Variants of Listeria Monocytogenes Using LC-MS/MS for Identification of Potential Surface Adhesins.
Tiong HK, Hartson SD, Muriana PM. Tiong HK, et al. Pathogens. 2016 May 17;5(2):40. doi: 10.3390/pathogens5020040. Pathogens. 2016. PMID: 27196934 Free PMC article. - A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation.
Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Li YH, et al. J Bacteriol. 2002 May;184(10):2699-708. doi: 10.1128/JB.184.10.2699-2708.2002. J Bacteriol. 2002. PMID: 11976299 Free PMC article. - Quorum sensing in group A Streptococcus.
Jimenez JC, Federle MJ. Jimenez JC, et al. Front Cell Infect Microbiol. 2014 Sep 12;4:127. doi: 10.3389/fcimb.2014.00127. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25309879 Free PMC article. Review. - Molecular mechanism of quorum sensing inhibition in Streptococcus by the phage protein paratox.
Rutbeek NR, Rezasoltani H, Patel TR, Khajehpour M, Prehna G. Rutbeek NR, et al. J Biol Chem. 2021 Sep;297(3):100992. doi: 10.1016/j.jbc.2021.100992. Epub 2021 Jul 21. J Biol Chem. 2021. PMID: 34298018 Free PMC article.
References
- Aaberge I S, Eng J, Lermark G, Løvik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–152. - PubMed
- Alloing G, Granadel C, Morrison D A, Claverys J P. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol Microbiol. 1996;21:471–478. - PubMed
- Alloing G, Martin B, Granadel C, Claverys J P. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum-sensing by the oligopeptide permease. Mol Microbiol. 1998;29:75–83. - PubMed
- Behr M A, Wilson M A, Gill W P, Salamon H, Skolnik G K, Rane S, Small P M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. - PubMed
- Campbell E A, Choi S Y, Masure H R. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol. 1998;27:929–939. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases