The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3 - PubMed (original) (raw)
The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3
G Peyroche et al. EMBO J. 2000.
Abstract
RNA polymerase I (Pol I) is dedicated to transcription of the large ribosomal DNA (rDNA). The mechanism of Pol I recruitment onto rDNA promoters is poorly understood. Here we present evidence that subunit A43 of Pol I interacts with transcription factor Rrn3: conditional mutations in A43 were found to disrupt the transcriptionally competent Pol I-Rrn3 complex, the two proteins formed a stable complex when co-expressed in Escherichia coli, overexpression of Rrn3 suppressed the mutant phenotype, and A43 and Rrn3 mutants showed synthetic lethality. Consistently, immunoelectron microscopy data showed that A43 and Rrn3 co-localize within the Pol I-Rrn3 complex. Rrn3 has several protein partners: a two-hybrid screen identified the C-terminus of subunit Rrn6 of the core factor as a Rrn3 contact, an interaction supported in vitro by affinity chromatography. Our results suggest that Rrn3 plays a central role in Pol I recruitment to rDNA promoters by bridging the enzyme to the core factor. The existence of mammalian orthologues of A43 and Rrn3 suggests evolutionary conservation of the molecular mechanisms underlying rDNA transcription in eukaryotes.
Figures
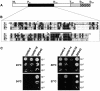
Fig. 1. Analysis of RPA43 mutants. (A) Schematic representation of the A43 subunit of S.cerevisiae. The black box represents the domain conserved in higher eukaryotes (see B). The grey box corresponds to the non-essential C-terminal part absent in the mutant protein encoded by the rpa43-14 allele (see C) and the hatched box to the hyperacidic C-terminal domain. (B) Sequence alignment of the conserved domain between putative homologues of the A43 subunit. S.c., Saccharomyces cerevisiae; C.a., Candida albicans; S.p., Schizosaccharomyces pombe; M.m., Mus musculus; H.s., Homo sapiens. Black boxes indicate the residues identical in at least three sequences. The black line localizes a 15 residue motif highly conserved from yeast to human. The asterisks indicate the three residues mutated in the protein encoded by the rpa43-6 allele. (C) Five rpa43 mutant alleles (rpa43-4, 6, 14, 18 and 24) borne on a centromeric plasmid were used to complement a chromosomal disruption of RPA43. Growth on YPD medium at different temperatures of serial dilutions of the resulting strains was analysed.
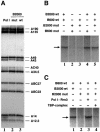
Fig. 2. A43-6 containing Pol I is inactive in specific transcription in vitro. (A) Subunit composition of Pol I contained in the B2000 fraction prepared from the rpa43-6 mutant (mut) and from a wild-type (wt) strain was analysed by western blotting using anti-Pol I antibodies. Purified RNA Pol I was used as a positive control. (B) Specific in vitro transcription of a mini 26S transcription unit was carried out with the B600 and B2000 fractions purified from the rpa43-6 mutant (mut) or a wild-type (wt) strain. B2000 fractions were normalized by their activity in a non-specific in vitro transcription assay. RNA was analysed by urea–PAGE followed by autoradiography. (C) Pol I–Rrn3 and TBP-containing complexes, purified from a wild-type strain, were added to the B2000mut fraction and specific in vitro transcription was performed as in (B).
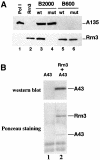
Fig. 3. Subunit A43 is an interaction target of Rrn3. (A) The Pol I–Rrn3 complex is unstable in the rpa43-6 mutant. The B2000 and B600 fractions purified from the rpa43-6 mutant (mut) and a wild-type (wt) strain were analysed for their Pol I and Rrn3 content by western blotting using antibodies directed to the A135 Pol I subunit or to Rrn3. The B2000 fractions were normalized by their activity in a non-specific in vitro transcription assay and identical amounts of proteins of the wild-type and mutant B600 fractions were used. Purified Pol I and Rrn3 are indicated. (B) Subunit A43 and Rrn3 form a stable binary complex. Escherichia coli extracts from cells expressing either subunit A43 (lane 1) or HA-(His)6-Rrn3 and subunit A43 (lane 2) were loaded onto a protein G–Sepharose column coated with a monoclonal antibody raised against the HA epitope. Proteins bound to the column were eluted by competition with the HA peptide. Eluted proteins were subjected to SDS–PAGE and the presence of A43 subunit was analysed by western blotting using polyclonal anti-A43 antibodies and by Ponceau Red staining.
Fig. 4. In vivo interactions between subunit A43 and Rrn3. (A) The rpa43-6 mutant strain was transformed with a 2µ plasmid carrying either the RRN3 or SPT15 gene (encoding TBP) or with a control vector. Growth of serial dilutions of the transformants at the permissive (24°C) and non-permissive (34°C) temperatures was monitored. (B) A centromeric plasmid carrying either RPA43 or rpa43-6 was introduced into GPy44 (rrn3-8, rpa43::LEU2, pNOY102) or D128 cells (RRN3, rpa43::LEU2, pNOY102). Growth of the double mutant strain (rrn3-8 rpa43-6), of the single mutant strains (rrn3-8 RPA43 and RRN3 rpa43-6) and of the wild-type strain was examined on galactose and glucose medium at 24°C.
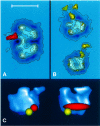
Fig. 5. Immunolocalization of subunit A43 and of the interaction site of Rrn3 in the Pol I three-dimensional structure. (A) Image analysis of RNA Pol I dimers labelled with anti-A43 polyclonal antibodies obtained upon analysis of 186 molecular images. The stain-excluding protein densities are outlined by contours of equal density. The difference image between the labelled and unlabelled molecules is shown in red where the contours correspond to positive differences at a threshold significance level of 5%. (B) Image analysis of RNA Pol I monomers labelled with anti-Rrn3 polyclonal antibodies. A total of 836 labelled RNA polymerase monomers was selected out of the original micrographs and 50.3% of the molecular images corresponded to two characteristic views: the –90° view, where the groove is located in the left part and the channel region in the lower left (upper); and the 0° view, where the groove is facing the observer and the channel region maps in the lower part (lower). The stain-excluding protein densities are outlined by contours of equal density. The significant differences between the labelled and unlabelled molecules are shown in yellow. (C) Surface representation of the three-dimensional RNA Pol I model showing the position of subunit A43 (red) and the interaction site of Rrn3 (yellow). The bar represents 20 nm in (A) and (B) and 12 nm in (C).
Fig. 6. Rrn3 interacts with Rrn6. (A) Two-hybrid interactions between Rrn3 and Rrn6. Strain Y190 was transformed with two plasmids, one allowing expression of Rrn3 fused to the Gal4 DBD, the other expression of Rrn6, Rrn6[772–894] or Rrn6[865–897] fused to the Gal4 AD. Activation of the lacZ and HIS3 reporter genes was monitored by staining the cells in the presence of XGal or replicating the cells on 3AT-containing medium, respectively. (B) The C-terminal domain of Rrn6 binds to immobilized Rrn3. Escherichia coli extracts containing (+) or not (–) recombinant HA-tagged Rrn3 were loaded onto protein G–Sepharose columns coated with 12CA5 anti-HA antibodies, then in vitro synthesized [35S]Rrn6[C1] (Load) were applied to the columns. Proteins eluted by competition with purified HA-peptide were subjected to SDS–PAGE and analysed by autoradiography. (C) Full-length Rrn3 binds to the immobilized C-terminal domain of Rrn6. Escherichia coli extracts containing GST (–) or GST–Rrn6[772–894] fusion protein (+) were bound to glutathione–Sepharose columns. After extensive washing, partially purified recombinant HA-tagged Rrn3 (Load) was chromatographed onto the columns. Proteins eluted with glutathione were subjected to SDS–PAGE and the presence of HA-Rrn3 analysed by western blotting using monoclonal anti-HA antibodies.
Similar articles
- The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits.
Peyroche G, Levillain E, Siaut M, Callebaut I, Schultz P, Sentenac A, Riva M, Carles C. Peyroche G, et al. Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):14670-5. doi: 10.1073/pnas.232580799. Epub 2002 Oct 29. Proc Natl Acad Sci U S A. 2002. PMID: 12407181 Free PMC article. - Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription.
Beckouet F, Labarre-Mariotte S, Albert B, Imazawa Y, Werner M, Gadal O, Nogi Y, Thuriaux P. Beckouet F, et al. Mol Cell Biol. 2008 Mar;28(5):1596-605. doi: 10.1128/MCB.01464-07. Epub 2007 Dec 17. Mol Cell Biol. 2008. PMID: 18086878 Free PMC article. - RNA polymerase I activation and hibernation: unique mechanisms for unique genes.
Fernández-Tornero C. Fernández-Tornero C. Transcription. 2018;9(4):248-254. doi: 10.1080/21541264.2017.1416267. Epub 2018 Jan 26. Transcription. 2018. PMID: 29372670 Free PMC article. Review. - The RNA polymerase I transcription machinery.
Russell J, Zomerdijk JC. Russell J, et al. Biochem Soc Symp. 2006;(73):203-16. doi: 10.1042/bss0730203. Biochem Soc Symp. 2006. PMID: 16626300 Free PMC article. Review.
Cited by
- RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing.
Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. Schneider DA, et al. Proc Natl Acad Sci U S A. 2006 Aug 22;103(34):12707-12. doi: 10.1073/pnas.0605686103. Epub 2006 Aug 14. Proc Natl Acad Sci U S A. 2006. PMID: 16908835 Free PMC article. - Site specific phosphorylation of yeast RNA polymerase I.
Gerber J, Reiter A, Steinbauer R, Jakob S, Kuhn CD, Cramer P, Griesenbeck J, Milkereit P, Tschochner H. Gerber J, et al. Nucleic Acids Res. 2008 Feb;36(3):793-802. doi: 10.1093/nar/gkm1093. Epub 2007 Dec 15. Nucleic Acids Res. 2008. PMID: 18084032 Free PMC article. - Evolution of eukaryotic transcription: insights from the genome of Giardia lamblia.
Best AA, Morrison HG, McArthur AG, Sogin ML, Olsen GJ. Best AA, et al. Genome Res. 2004 Aug;14(8):1537-47. doi: 10.1101/gr.2256604. Genome Res. 2004. PMID: 15289474 Free PMC article. - Structure of the human RNA polymerase I elongation complex.
Zhao D, Liu W, Chen K, Wu Z, Yang H, Xu Y. Zhao D, et al. Cell Discov. 2021 Oct 20;7(1):97. doi: 10.1038/s41421-021-00335-5. Cell Discov. 2021. PMID: 34671025 Free PMC article. - Inositol pyrophosphates regulate RNA polymerase I-mediated rRNA transcription in Saccharomyces cerevisiae.
Thota SG, Unnikannan CP, Thampatty SR, Manorama R, Bhandari R. Thota SG, et al. Biochem J. 2015 Feb 15;466(1):105-14. doi: 10.1042/BJ20140798. Biochem J. 2015. PMID: 25423617 Free PMC article.
References
- Bateman E. and Paule,M.R. (1986) Regulation of eucaryotic ribosomal RNA transcription by RNA polymerase modification. Cell, 47, 445–450. - PubMed
- Bell S.P., Learned,R.M., Jantzen,H.M. and Tjian,R. (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science, 241, 1192–1197. - PubMed
- Bréant B., Buhler,J.-M., Sentenac,A. and Fromageot,P. (1983) On the phosphorylation of yeast RNA polymerases A and B. Eur. J. Biochem., 130, 247–251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases