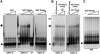She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p - PubMed (original) (raw)
She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p
F Böhl et al. EMBO J. 2000.
Abstract
RNA localization is a widespread mechanism to achieve localized protein synthesis. In budding yeast, localization of ASH1 mRNA controls daughter cell-specific accumulation of the transcriptional regulator Ash1p, which determines mating type switching. ASH1 mRNA localization depends on four independently acting sequences ('zipcodes') within the mRNA. In addition, the class V myosin Myo4p and a set of She proteins with as yet unknown function are essential for ASH1 localization. Here we show that She2p is a novel RNA-binding protein that binds specifically to ASH1 mRNA in vivo and to ASH1 RNA zip codes in vitro. She2p can interact with She3 protein via She3p's C-terminus and becomes localized to the daughter cell tip upon ASH1 expression. The N-terminal coiled-coil domain of She3p is required to form an RNA-independent complex with the heavy chain of the myosin motor protein Myo4p. She2p and She3p are the first examples of adapters for tethering a localized mRNA to the motor protein and might serve as prototypes for RNA-motor protein adapters.
Figures
Fig. 1. She3p binds to the motor Myo4p and to She2p via different domains. (A) Schematic representation of the constructs used in the in vivo and in vitro interaction assays. Numbers below boxes correspond to amino acid position. Hatched boxes in Myo4p and She3p represent predicted coiled-coil regions. (B) Two hybrid interaction analysis. She2-, Myo4 tail- and Myo2 tail-Gal4 DNA binding domain fusions (‘bait’) were coexpressed with Gal4 activation domain alone or fusions (‘prey’) to Myo4p tail, She3p, She3p N-terminus and She3p C-terminus. Interaction was indicated by growth on medium lacking histidine. Whereas She2p showed interaction with full-length She3p and the She3p C-terminus, Myo4p tail interacted with She3p and She3p N-terminus. (C) In vitro interaction of She3p with Myo4p tail and She2p. Recombinant GST fusions of She2p, Myo4p tail, a Myo4 tail lacking the predicted coiled-coil domain or GST alone were immobilized to glutathione–Sepharose beads and incubated with in vitro translated 35S-labelled She3p (double arrows). Bound She3p was eluted, and applied to an SDS–PAGE gel together with one-fifth of the in vitro translated She3p used for pulldown (‘input’). Numbers indicate positions of molecular weight markers (in kDa). The band seen at 40 kDa is a degradation product of She3p. (D) In vitro interaction of She2p with the C-terminus of She3p. In vitro translated domains of She3p were affinity purified with GST–She2p beads as described above. Only the She3p C-terminal domain bound to GST–She2p.
Fig. 2. She3p association with Myo4p in vivo is RNA independent. (A) Western blot analysis of a 5–25% sucrose gradient fractionation of a cell extract from strain RJY758 (MYO4–HA6, SHE3-TEVProtA, YEplac181-ASH1) after treatment with RNase A (upper panels) or mock treatment (lower panels). Numbers on top of the panels indicate fraction number (1 = top), numbers on the bottom indicate migration behaviour (in Svedberg units) of protein standards in a parallel gradient. (B) Immunoprecipitation of She3–ProtA by IgG–agarose beads from peak fractions of the RNase A-treated extracts reveals coprecipitation of Myo4p. Upper panel: western blot analysis using anti-HA antibody 12CA5; lower panel: western blot analysis using peroxidase–anti-peroxidase complex to detect She3–protein A fusion.
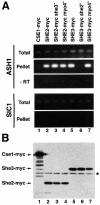
Fig. 3. Specific coprecipitation of ASH1 mRNA with myc-tagged She3p and She2p. Myc-tagged proteins from cell extracts of strains containing or lacking functional SHE genes were precipiated with anti-myc antibody 9E11 coupled to magnetic beads essentially as described in Münchow et al. (1999). Eluted material was prepared for RT–PCR or western blot analysis. (A) RT–PCR analysis. A 247 bp fragment covering part of the ASH1 3′UTR or a 316 bp fragment covering the 3′ part of SIC1 mRNA were amplified from total extract and immunoprecipitates of myc-tagged proteins. No PCR product was observed when reverse transcriptase was omitted (‘–RT’). (B) To show equal efficiency of immunoprecipitations in myc-tagged strains, identical aliquots of the immunoprecipitates were separated on 10% SDS–PAGE and myc-tagged proteins detected by western blotting using anti-myc antibody 9E10. Lane 1, Cse1p–myc; lanes 2–4, She2p–myc; lanes 5–7, She3–myc. An asterisk marks the position of the antibody heavy chain.
Fig. 4. Purified GST–She2p binds ASH1 RNA in vitro through the localization elements E1–E3. (A) Overview of the results of _ASH1_–GST–She2p pulldown experiments. At the top of the panel ASH1 mRNA is sketched with the positions of the localization elements E1, E2A, E2B and E3 (hatched bars). Numbers below indicate nucleotide position (AUG = 1). Arrows indicate the start and end of the RNAs used in the pulldown experiments. Numbers in parentheses on the left indicate the start and end of the RNAs on the nucleotide level. The right column indicates relative recovery of input RNA on the glutathione beads. +++, recovery >50%; ++, >35%; +, >15%; –, <10%. Results are mean values of three to five independent experiments. (B) Representative denaturing polyacrylamide gel showing specific precipitation of 32P-labelled localization elements E2B, E3 and ASH1-U by GST–She2p (lanes 6–8) but not of the mutant U* or an RNA lacking localization elements (‘inter’, lanes 9 and 10). No binding of RNA was observed when beads where loaded with GST (lanes 11–15). Lanes 1–5 show input RNAs. (C) Schematic overview of predicted secondary structures of localization elements E3, ASH1-U and U*. The arrow indicates disrupted doubled-stranded RNA stem in mutant element U*.
Fig. 5. She2p binding to ASH1-U is specific and enhanced by She3p. Probes used are indicated at the bottom of each panel, proteins at the top. (A) RNA mobility shift assay using 32P-labelled probes of ASH1-U or mutant U* and increasing concentrations of GST, GST–She2p (2.5, 5.0, 7.5, 10, 20, 40 ng/µl) or GST–She3p (10, 20, 30, 40, 60 ng/µl). The arrowhead indicates the position of free probe and the black arrow the position of an ASH1-U RNA complex with GST–She2p that cannot be detected with GST–She3p or mutant U*. (B) Mobility shift using constant GST–She2p (2.5 ng/µl) and increasing concentrations of GST or GST–She3p (10, 20, 30, 40, 60 ng/µl). Increasing the amount of She3p but not of GST enhances She2p–ASH1-U complex formation (black arrow) and leads to the formation of an RNA–protein complex with lower mobility (open arrow). (C) Mobility shift assay using the IRE as probe. No complex formation is observed.
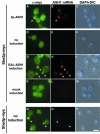
Fig. 6. She2p–myc9 localizes to the bud tip upon ASH1 expression. Overexpression of ASH1 mRNA from a high-copy plasmid (A–C) or from the heterologous GAL1 promoter (G–H) induces a shift of She2p localization to the bud tip (arrowheads). Localization of She3–myc6 to the bud tip is independent of ASH1 expression (N–P). A combination of indirect immunofluorescence against myc-tagged proteins and in situ hybridization against ASH1 mRNA was performed as described in Materials and methods. Immunofluorescence staining against She2p–myc9 is shown in (A), (D), (G) and (K), whereas staining in (N) reflects She3p–myc6 localization. (B), (E), (H), (L) and (O) show localization of ASH1 mRNA by in situ hybridization using CY3-labelled ASH1 antisense oligos. (C), (F), (I), (M) and (P) are composite images of differential interference contrast (DIC) and DNA staining by DAPI. Bar, 5 µm.
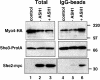
Fig. 7. Coprecipitation of She2–myc with She3–ProtA and Myo4_–HA_ upon ASH1 RNA overexpression. Western blot analysis of cell extracts from strains RJY750 (SHE3–ProtA, MYO4–HA6, ‘control’), RJY756 (SHE3–ProtA, MYO4–HA6, SHE2–myc3, pGAL1; ‘–ASH1’) and RJY757 (SHE3–ProtA, MOY4–HA6, SHE2–myc3, pGAL1-ASH1; ‘+ASH1’), and corresponding pellets from an anti-protein A precipitation (‘IgG-beads’). The blot was cut into three pieces and probed with antibodies against HA, myc or protein A. Numbers on the right indicate the positions of size markers (in kDa).
Similar articles
- The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p.
Takizawa PA, Vale RD. Takizawa PA, et al. Proc Natl Acad Sci U S A. 2000 May 9;97(10):5273-8. doi: 10.1073/pnas.080585897. Proc Natl Acad Sci U S A. 2000. PMID: 10792032 Free PMC article. - She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA.
Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. Long RM, et al. EMBO J. 2000 Dec 1;19(23):6592-601. doi: 10.1093/emboj/19.23.6592. EMBO J. 2000. PMID: 11101531 Free PMC article. - RNA-protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex.
Gonsalvez GB, Lehmann KA, Ho DK, Stanitsa ES, Williamson JR, Long RM. Gonsalvez GB, et al. RNA. 2003 Nov;9(11):1383-99. doi: 10.1261/rna.5120803. RNA. 2003. PMID: 14561888 Free PMC article. - RNA localization: SHEdding light on the RNA-motor linkage.
Kwon S, Schnapp BJ. Kwon S, et al. Curr Biol. 2001 Mar 6;11(5):R166-8. doi: 10.1016/s0960-9822(01)00084-7. Curr Biol. 2001. PMID: 11267883 Review. - RNA asymmetric distribution and daughter/mother differentiation in yeast.
Darzacq X, Powrie E, Gu W, Singer RH, Zenklusen D. Darzacq X, et al. Curr Opin Microbiol. 2003 Dec;6(6):614-20. doi: 10.1016/j.mib.2003.10.005. Curr Opin Microbiol. 2003. PMID: 14662358 Free PMC article. Review.
Cited by
- Widespread mRNA association with cytoskeletal motor proteins and identification and dynamics of myosin-associated mRNAs in S. cerevisiae.
Casolari JM, Thompson MA, Salzman J, Champion LM, Moerner WE, Brown PO. Casolari JM, et al. PLoS One. 2012;7(2):e31912. doi: 10.1371/journal.pone.0031912. Epub 2012 Feb 16. PLoS One. 2012. PMID: 22359641 Free PMC article. - Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors.
Bretscher A. Bretscher A. J Cell Biol. 2003 Mar 17;160(6):811-6. doi: 10.1083/jcb.200301035. J Cell Biol. 2003. PMID: 12642608 Free PMC article. Review. - Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA.
Du TG, Jellbauer S, Müller M, Schmid M, Niessing D, Jansen RP. Du TG, et al. EMBO Rep. 2008 Aug;9(8):781-7. doi: 10.1038/embor.2008.112. Epub 2008 Jun 20. EMBO Rep. 2008. PMID: 18566598 Free PMC article. - Distinct roles for Khd1p in the localization and expression of bud-localized mRNAs in yeast.
Hasegawa Y, Irie K, Gerber AP. Hasegawa Y, et al. RNA. 2008 Nov;14(11):2333-47. doi: 10.1261/rna.1016508. Epub 2008 Sep 19. RNA. 2008. PMID: 18805955 Free PMC article. - Role of Loc1p in assembly and reorganization of nuclear ASH1 messenger ribonucleoprotein particles in yeast.
Niedner A, Müller M, Moorthy BT, Jansen RP, Niessing D. Niedner A, et al. Proc Natl Acad Sci U S A. 2013 Dec 24;110(52):E5049-58. doi: 10.1073/pnas.1315289111. Epub 2013 Dec 9. Proc Natl Acad Sci U S A. 2013. PMID: 24324176 Free PMC article.
References
- Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Adinolfi S., Bagni,C., Castiglione Morelli,M.A., Fraternali,F., Musco,G. and Pastore,A. (1999) Novel RNA-binding motif: The KH module. Biopolymers, 51, 153–164. - PubMed
- Bashirulla A., Cooperstock,R.L. and Lipshitz,H.D. (1998) RNA localization in development. Annu. Rev. Biochem., 67, 335–394. - PubMed
- Bassell G.J., Oleynikov,Y. and Singer,R.H. (1999) The travels of mRNAs through all cells large and small. FASEB J., 13, 447–454. - PubMed
- Bertrand E., Chartrand,P., Schaefer,M., Shenoy,S.M., Singer,R.H. and Long,R.M. (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell, 2, 437–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases