Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus - PubMed (original) (raw)
Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus
J A Morrissey et al. Infect Immun. 2000 Nov.
Abstract
From a mass-excised Staphylococcus aureus lambdaZapII expression library, we cloned an operon encoding a novel ABC transporter with significant homology to bacterial siderophore transporter systems. The operon encodes four genes designated sstA, -B, -C, and -D encoding two putative cytoplasmic membrane proteins (sstA and sstB), an ATPase (sstC), and a membrane-bound 38-kDa lipoprotein (sstD). The sst operon is preceded by two putative Fur boxes, which indicated that expression of the sst operon was likely to be iron dependent. SstD was overexpressed in Escherichia coli, purified by Triton X-114 phase partitioning, and used to generate monospecific antisera in rats. Immunoblotting studies located SstD in the membrane fraction of S. aureus and showed that expression of the lipoprotein was reduced under iron-rich growth conditions. Triton X-114 partitioning studies on isolated membranes provided additional biochemical evidence that SstD in S. aureus is a lipoprotein. Immunoreactive polypeptides of approximately 38 kDa were detected in a wide range of staphylococcal species, but no antigenic homolog was detected in Bacillus subtilis. Expression of SstD in vivo was confirmed by immunoblotting studies with S. aureus recovered from a rat intraperitoneal chamber implant model. To further define the contribution of SstD in promoting growth of S. aureus in vitro and in vivo, we used antisense RNA technology to modulate expression of SstD. Expression of antisense sstD RNA in S. aureus resulted in a decrease in SstD expression under both iron-rich and iron-restricted growth conditions. However, this reduction in SstD levels did not affect the growth of S. aureus in vitro in an iron-limited growth medium or when grown in an intraperitoneal rat chamber implant model in vivo.
Figures
FIG. 1
(A) Southern blot analysis showing DNA homology among the S. aureus BB sstABCD operon, other_S. aureus_ strains, and S. epidermidis. Plasmid and genomic DNAs were digested with Mun_I, and the Southern blots were hybridized with a digoxigenin-labeled PCR product amplified from BB genomic DNA using primers 38A for and 38B rev. Lanes: M, λ_Hin_dIII/phiX174 Hae_III DNA markers; 1, S. epidermidis 901; 2, S. aureus BB; 3, S. aureus W; 4, S. aureus RN4220; 5, plasmid pJM10; 6, diluted PCR product used as the probe. (B) Organization of the S. aureus sstABCD ABC transporter operon. (C) The upstream sequences of the sstABCD operon showing one of the putative Fur box sequences in bold and the position of the potential inverted repeat region. The ribosome-binding site and the translational start site are underlined. (D) Alignment of the_sstABCD putative Fur box sequences with the E. coli Fur box consensus sequence (9) and the Fur box sequences from the S. aureus sirA (14) and_fhuA (36) genes. The differences are highlighted.
FIG. 2
Immunoblots with rat anti-SstD serum showing membrane association, iron regulation, and Triton X-114 solubility of SstD. (A) Membrane fractions of S. aureus BB grown for 6 h under iron-restricted (lane 1) or iron-rich (lane 2) conditions. The 38-kDa SstD lipoprotein is indicated. (B) Triton X-114 extracts of S. aureus BB grown for 18 h in RPMI 1640 medium (lane 2) and_S. aureus_ BB (lane 3) or S. epidermidis 901 (lane 4) grown for 6 h in CRPMI. Lane 1 shows the membrane fraction of S. aureus BB grown for 6 h in CRPMI. The 38-kDa SstD protein is indicated.
FIG. 3
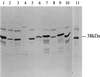
Immunoblot of Triton-X114 extracts prepared from_S. aureus_, coagulase-negative staphylococci, and B. subtilis grown under iron-restricted conditions in vitro and_S. aureus_ BB grown in vivo. The immunoblots were reacted with monospecific rat antiserum to the 38-kDa SstD lipoprotein. Lanes: 1, S. aureus BB; 2, S. haemolyticus; 3, S. carnosus; 4, B. subtilis; 5, S. hominis; 6, S. warnerii; 7,S. cohni, 8, S. lugdunensis; 9, S. saprophyticus; 10, S. epidermidis; 11, S. aureus BB grown in vivo.
FIG. 4
(A) Northern blot analysis of antisense sstD_transcripts. Total RNAs were isolated from logarithmically growing_S. aureus strains grown under iron-rich or iron-restricted conditions in vitro. The Northern blot was hybridized with digoxigenin-labeled sstD RNA. The approximately 1-kb antisense transcript is indicated. (B) Western blot analysis of membrane fractions from S. aureus RN6390-B showing the decrease in SstD protein production in the presence of _sstD_antisense RNA. Lanes: 1, RN6390-B (with Fe); 2, RN6390-B (without Fe); 3, RN6390-B pS10 (with Fe); 4, RN6390-B pS10 (without Fe); 5, RN6390-B pS1038 (with Fe); 6, RN6390-B pS1038 (without Fe). (C) Immunoblot of membrane preparations from S. aureus recovered without subculture from the chambers and probed with anti-SstD serum. Lanes: 1, RN6390-B S1038; 2, RN6390-B S10.
Similar articles
- Conservation, surface exposure, and in vivo expression of the Frp family of iron-regulated cell wall proteins in Staphylococcus aureus.
Morrissey JA, Cockayne A, Hammacott J, Bishop K, Denman-Johnson A, Hill PJ, Williams P. Morrissey JA, et al. Infect Immun. 2002 May;70(5):2399-407. doi: 10.1128/IAI.70.5.2399-2407.2002. Infect Immun. 2002. PMID: 11953376 Free PMC article. - Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter.
Cockayne A, Hill PJ, Powell NB, Bishop K, Sims C, Williams P. Cockayne A, et al. Infect Immun. 1998 Aug;66(8):3767-74. doi: 10.1128/IAI.66.8.3767-3774.1998. Infect Immun. 1998. PMID: 9673260 Free PMC article. - Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus.
Beasley FC, Vinés ED, Grigg JC, Zheng Q, Liu S, Lajoie GA, Murphy ME, Heinrichs DE. Beasley FC, et al. Mol Microbiol. 2009 May;72(4):947-63. doi: 10.1111/j.1365-2958.2009.06698.x. Epub 2009 Apr 14. Mol Microbiol. 2009. PMID: 19400778 - Involvement of SirABC in iron-siderophore import in Staphylococcus aureus.
Dale SE, Sebulsky MT, Heinrichs DE. Dale SE, et al. J Bacteriol. 2004 Dec;186(24):8356-62. doi: 10.1128/JB.186.24.8356-8362.2004. J Bacteriol. 2004. PMID: 15576785 Free PMC article. - Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus.
Xiong A, Singh VK, Cabrera G, Jayaswal RK. Xiong A, et al. Microbiology (Reading). 2000 Mar;146 ( Pt 3):659-668. doi: 10.1099/00221287-146-3-659. Microbiology (Reading). 2000. PMID: 10746769
Cited by
- High-affinity binding of the staphylococcal HarA protein to haptoglobin and hemoglobin involves a domain with an antiparallel eight-stranded beta-barrel fold.
Dryla A, Hoffmann B, Gelbmann D, Giefing C, Hanner M, Meinke A, Anderson AS, Koppensteiner W, Konrat R, von Gabain A, Nagy E. Dryla A, et al. J Bacteriol. 2007 Jan;189(1):254-64. doi: 10.1128/JB.01366-06. Epub 2006 Oct 13. J Bacteriol. 2007. PMID: 17041047 Free PMC article. - Conservation, surface exposure, and in vivo expression of the Frp family of iron-regulated cell wall proteins in Staphylococcus aureus.
Morrissey JA, Cockayne A, Hammacott J, Bishop K, Denman-Johnson A, Hill PJ, Williams P. Morrissey JA, et al. Infect Immun. 2002 May;70(5):2399-407. doi: 10.1128/IAI.70.5.2399-2407.2002. Infect Immun. 2002. PMID: 11953376 Free PMC article. - Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors.
Heilbronner S, Holden MT, van Tonder A, Geoghegan JA, Foster TJ, Parkhill J, Bentley SD. Heilbronner S, et al. FEMS Microbiol Lett. 2011 Sep;322(1):60-7. doi: 10.1111/j.1574-6968.2011.02339.x. Epub 2011 Jul 4. FEMS Microbiol Lett. 2011. PMID: 21682763 Free PMC article. - Global network analysis of drug tolerance, mode of action and virulence in methicillin-resistant S. aureus.
Overton IM, Graham S, Gould KA, Hinds J, Botting CH, Shirran S, Barton GJ, Coote PJ. Overton IM, et al. BMC Syst Biol. 2011 May 12;5:68. doi: 10.1186/1752-0509-5-68. BMC Syst Biol. 2011. PMID: 21569391 Free PMC article. - Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing.
Doherty N, Holden MT, Qazi SN, Williams P, Winzer K. Doherty N, et al. J Bacteriol. 2006 Apr;188(8):2885-97. doi: 10.1128/JB.188.8.2885-2897.2006. J Bacteriol. 2006. PMID: 16585750 Free PMC article.
References
- Arbuthnott J P, Arbuthnott E, Arbuthnott A D J, Pike W J, Cockayne A. Investigation of microbial growth in vivo: evaluation of a novel in vivochamber implant system. FEMS Microbiol Lett. 1992;100:75–80. - PubMed
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1994.
- Brusca J S, Radolf J D. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 1994;228:182–193. - PubMed
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtiliscontains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. - PubMed
- Chang S, Cohen N. High frequency transformation of Bacillus subtilisprotoplasts by plasmid DNA. Mol Gen Genet. 1979;168:111–115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases