Characterization of the signal that directs Tom20 to the mitochondrial outer membrane - PubMed (original) (raw)
Characterization of the signal that directs Tom20 to the mitochondrial outer membrane
S Kanaji et al. J Cell Biol. 2000.
Abstract
Tom20 is a major receptor of the mitochondrial preprotein translocation system and is bound to the outer membrane through the NH(2)-terminal transmembrane domain (TMD) in an Nin-Ccyt orientation. We analyzed the mitochondria-targeting signal of rat Tom20 (rTom20) in COS-7 cells, using green fluorescent protein (GFP) as the reporter by systematically introducing deletions or mutations into the TMD or the flanking regions. Moderate TMD hydrophobicity and a net positive charge within five residues of the COOH-terminal flanking region were both critical for mitochondria targeting. Constructs without net positive charges within the flanking region, as well as those with high TMD hydrophobicity, were targeted to the ER-Golgi compartments. Intracellular localization of rTom20-GFP fusions, determined by fluorescence microscopy, was further verified by cell fractionation. The signal recognition particle (SRP)-induced translation arrest and photo-cross-linking demonstrated that SRP recognized the TMD of rTom20-GFP, but with reduced affinity, while the positive charge at the COOH-terminal flanking segment inhibited the translation arrest. The mitochondria-targeting signal identified in vivo also functioned in the in vitro system. We conclude that NH(2)-terminal TMD with a moderate hydrophobicity and a net positive charge in the COOH-terminal flanking region function as the mitochondria-targeting signal of the outer membrane proteins, evading SRP-dependent ER targeting.
Figures
Figure 1
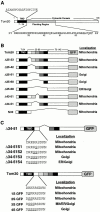
Schematic presentation of rTom20 and the flanking region-deleted mutants of rTom22-GFP fusions. (A) Schematic structure of rTom20. Amino acid sequences of the TMD and the flanking region are shown. (B) Schematic structure of the flanking region–deleted rTom20-GFP fusions. (C) Structure of the constructs carrying mutations in the COOH-terminal flanking region.
Figure 2
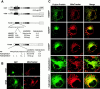
Expression of wild-type rTom20-GFP in COS-7 cells. COS-7 cells were transfected with the expression vector containing the rTom20-GFP cDNA as described in Materials and Methods. The cells were incubated with MitoTracker, and fluorescent images of GFP (green) and MitoTracker (red) were taken by a confocal microscope.
Figure 3
Basic amino acids at the COOH-terminal flanking region of the TMD are critical for targeting rTom20-GFP fusions to mitochondria. COS-7 cells were transfected with the indicated constructs in the expression vectors. The cells were costained with MitoTracker. Other conditions were the same as in Fig. 2. (A) Localization of rTom20-GFP constructs carrying mutations in the COOH-terminal flanking region. Merged fluorescent images of GFP and MitoTracker are shown. (B) Fluorescent images of MitoTracker and rTom20-GFP mutants in which the basic amino acids in the flanking region were replaced by serine residues.
Figure 4
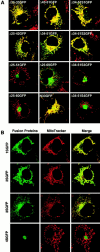
The rTom20-GFP constructs that are incompetent for mitochondria-targeting localized to the ER-Golgi compartments. (A) Localization of Δ25-60GFP to the ER-Golgi compartments. COS-7 cells expressing Δ25-60GFP were immunostained with IgGs against GM130 (Golgi marker) or Calnexin (ER marker). Fluorescent images of GFP (green), GM130 (red), and Calnexin (red) were taken by a confocal microscope. (B) Effect of nocodazole or brefeldin A (BFA) on intracellular localization of Δ25-60GFP. Δ25-69GFP-expressing COS-7 cells were treated with 20 μg/ml nocodazole for 2 h or with 5 μg/ml BFA for 1 h. The cells were then stained with MitoTracker. In a different experiment, Δ25-69GFP-expressing COS-7 cells were treated with BFA as described above, and then immunostained with IgGs against Calnexin (red).
Figure 5
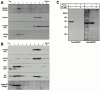
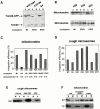
Subcellular localization of rTom20-GFP constructs as examined by discontinuous sucrose gradient centrifugation. (A) Localization of Δ25-69GFP to the ER-Golgi compartments. rTom20-GFP– or Δ25-69GFP–transfected COS-7 cells were subfractionated by sucrose density gradient as described in Materials and Methods. GM130 (Golgi marker), rTom20, rTom20-GFP, and Δ25-69GFP in each fraction were detected by Western blotting. (B) Subcellular localization of Δ25-33GFP, H22GFP, and 3SGFP. Δ25-33GFP-, H22GFP-, or 3SGFP-transfected COS-7 cells were subfractionated by sucrose density gradient centrifugation. The Golgi-enriched fractions were detected using anti–Golgin97 antibodies. Other conditions were the same as those described in A. (C) Membrane topology of rTom20-GFP and Δ25-69GFP expressed in COS-7 cells. Fractions 3 and 8 in A were treated with 100 μg/ml proteinase K for 30 min at 0°C in the presence or absence of 1% Triton X-100. The reaction mixtures were analyzed by SDS-PAGE and immunoblotting with anti–rTom20 IgG.
Figure 6
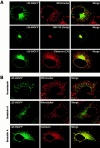
TMD length or hydrophobicity is a critical determinant for mitochondria localization. (A) Schematic presentation of rTom20-GFP constructs in which the NH2-terminal hydrophilic segment and the TMD were replaced by those of Syt II (ST27 and ST20), leucine repeats were inserted into the TMD (H20, H22 and H24), or glycine or tyrosine residues in the TMD were replaced by leucine residues (Hydrophobic-1 and -2). Hydrophobicity, hydropathic index/residue; Mit, mitochondria; PM, plasma membrane. (B) Localization of ST27 and ST20 in COS-7 cells. Confocal images of GFP (green) and MitoTracker (red) are shown. (C) Mitochondria localization of rTom20-GFP fusions depends on the TMD length or hydrophobicity. The indicated rTom20-GFP constructs were expressed in COS-7 cells. The cells were costained with MitoTracker and fluorescence images were taken by a confocal microscope.
Figure 7
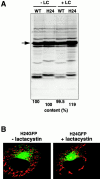
Proteasome inhibitor lactacystin does not affect the expression level and subcellular localization of rTom20-GFP constructs in COS-7 cells. (A) Effect of lactacystin on the expression of rTom20-GFP and H24GFP in COS-7 cells. COS-7 cells transfected with wild-type rTom20-GFP (WT) or H24GFP were grown at 37°C for 24 h, which were then incubated with or without 20 μM lactacystin (LC) for 10 h at 37°C. The cell lysates were subjected to SDS-PAGE followed by Western blotting with anti–rTom20 IgG. WT and H24 proteins are shown by an arrow. (B) Effect of lactacystin on the localization of H24GFP in COS-7 cells. H24GFP-expressed COS-7 cells prepared as in A were stained with MitoTracker. Merged confocal images of GFP (green) and MitoTracker (red) are shown.
Figure 8
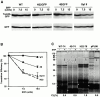
Insertion of rTom-GFP fusion proteins into rat liver mitochondria or dog pancreas microsomes in vitro. (A) Post-translational insertion of rTom20-GFP fusions into mitochondria. Wild-type rTom20-GFP, Δ25-69GFP, and Δ34-51S4GFP were synthesized in vitro and subjected to mitochondrial import. The reaction mixtures were subjected to sodium carbonate, pH 11.5, extraction. The mitochondria (M) and supernatant (S) fractions were analyzed by SDS-PAGE and subsequent Western blotting with anti–rTom20 IgG. Endogenous Tom20 is also shown. Mt, mitochondria; ER/G, ER-Golgi compartments. (B) Post-translational insertion into mitochondria and cotranslational insertion into the ER exhibit distinct dependency upon the TMD length. In vitro–synthesized H20GFP, H22GFP, and H24GFP were subjected to post-translational mitochondrial import and cotranslational ER import. Other conditions were the same as those described in A. The efficiency of post-translational mitochondria-insertion (C) and cotranslational ER-integration (D) of various rTom20-GFP constructs was quantified from the data in A and B. Integration efficiency (%) = [signal in M/ signal in (M + S)] × 100. (E) Insertion of nascent H24GFP into microsomal membrane. Truncated H24GFP (202 residues) was synthesized in the reticulocyte lysate system at 28°C for 25 min. After addition of 2 mM cycloheximide, the reaction mixture was divided into three aliquots. Two aliquots were incubated with microsomes in the absence (Control) or presence of 25 mM GMP-PNP (GMP-PNP) at 28°C for 20 min. The other aliquot was incubated with 10 mM NEM at 0°C for 10 min, followed by incubation with microsomes at 28°C for 20 min. All the reaction mixtures were subjected to the alkali floatation to separate the membrane (M) and unfloated (S) fractions. Both fractions were subjected to SDS-PAGE followed by Western blotting using anti–rTom20 IgG. (F) Insertion of truncated rTom20-GFP or 7SGFP into mitochondria. Truncated wild-type rTom20-GFP (WT) or 7SGFP (202 residues each) was synthesized in the reticulocyte lysate at 28°C for 25 min and divided into three aliquots. Two aliquots were incubated with mitochondria in the presence of 2 mM cycloheximide (CHI) or 2 mM puromycin plus 25 mM EDTA at 28°C for 60 min. The other aliquot was incubated with mitochondria at 28°C for 40 min, and then incubated in the presence of puromycin plus EDTA for 20 min (chase). Other conditions are as described in E.
Figure 9
Interaction of the nascent rTom20-GFP constructs with SRP. (A) Translation arrest of rTom20-GFP constructs. mRNA for wild-type rTom20-GFP (WT), H22GFP, 4SGFP, or Syt II was cotranslated with the control GFP mRNA in the wheat germ lysate at 30°C for 40 min in the presence or absence of the indicated amounts of SRP. Translation products were analyzed by SDS-PAGE and subsequent Western blotting using anti–rTom20 IgG, anti–GFP IgG, or anti–Syt II IgG. (B) The Western blot images were analyzed by LAS1000 (Fuji Film Co.). Intensities of bands of the translated rTom20-GFP constructs were normalized by those of GFP, and then the translation efficiencies were calculated taking those in the absence of SRP as 100%. (C) Interaction of SRP with the nascent chains of rTom20-GFP constructs and preprolactin (pPL) as examined by photo–cross-linking. The truncated mRNAs indicated were translated in the wheat germ lysate system in the presence of 35S-methionine, TDBA-lys-tRNA, and, where indicated, SRP at 25°C for 20 min. After addition of 2 mM cycloheximide, the reaction mixtures were UV irradiated, and then subjected to SDS-PAGE and autoradiography. Arrowheads, cross-linked products; asterisks, truncated peptides synthesized. CL (%), efficiency of cross-linking = [signal in arrowhead/signal in asterisk] × 100.
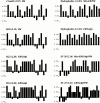
Figure 10
Hydrophobicity plots of TMDs of the rTom20-GFP constructs. The algorithm of Kyte and Doolittle was used with a window size of one amino acid residue. The average hydrophobicity is shown in parentheses.
Similar articles
- Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane.
Horie C, Suzuki H, Sakaguchi M, Mihara K. Horie C, et al. Mol Biol Cell. 2002 May;13(5):1615-25. doi: 10.1091/mbc.01-12-0570. Mol Biol Cell. 2002. PMID: 12006657 Free PMC article. - Targeting and assembly of rat mitochondrial translocase of outer membrane 22 (TOM22) into the TOM complex.
Nakamura Y, Suzuki H, Sakaguchi M, Mihara K. Nakamura Y, et al. J Biol Chem. 2004 May 14;279(20):21223-32. doi: 10.1074/jbc.M314156200. Epub 2004 Feb 25. J Biol Chem. 2004. PMID: 14985332 - Novel targeting signals mediate the sorting of different isoforms of the tail-anchored membrane protein cytochrome b5 to either endoplasmic reticulum or mitochondria.
Hwang YT, Pelitire SM, Henderson MP, Andrews DW, Dyer JM, Mullen RT. Hwang YT, et al. Plant Cell. 2004 Nov;16(11):3002-19. doi: 10.1105/tpc.104.026039. Epub 2004 Oct 14. Plant Cell. 2004. PMID: 15486098 Free PMC article. - Protein translocation across and integration into membranes.
Rapoport TA. Rapoport TA. CRC Crit Rev Biochem. 1986;20(1):73-137. doi: 10.3109/10409238609115901. CRC Crit Rev Biochem. 1986. PMID: 3007024 Review. - Common principles of protein translocation across membranes.
Schatz G, Dobberstein B. Schatz G, et al. Science. 1996 Mar 15;271(5255):1519-26. doi: 10.1126/science.271.5255.1519. Science. 1996. PMID: 8599107 Review.
Cited by
- A single-component, light-assisted uncaging switch for endoproteolytic release.
Cui M, Lee S, Ban SH, Ryu JR, Shen M, Yang SH, Kim JY, Choi SK, Han J, Kim Y, Han K, Lee D, Sun W, Kwon HB, Lee D. Cui M, et al. Nat Chem Biol. 2024 Mar;20(3):353-364. doi: 10.1038/s41589-023-01480-6. Epub 2023 Nov 16. Nat Chem Biol. 2024. PMID: 37973890 - Go your own way: membrane-targeting sequences.
Wojcik S, Kriechbaumer V. Wojcik S, et al. Plant Physiol. 2021 Apr 2;185(3):608-618. doi: 10.1093/plphys/kiaa058. Plant Physiol. 2021. PMID: 33822216 Free PMC article. - Par6 alpha interacts with the dynactin subunit p150 Glued and is a critical regulator of centrosomal protein recruitment.
Kodani A, Tonthat V, Wu B, Sütterlin C. Kodani A, et al. Mol Biol Cell. 2010 Oct 1;21(19):3376-85. doi: 10.1091/mbc.E10-05-0430. Epub 2010 Aug 18. Mol Biol Cell. 2010. PMID: 20719959 Free PMC article. - The Functional Characterization of GCaMP3.0 Variants Specifically Targeted to Subcellular Domains.
Kempmann A, Gensch T, Offenhäusser A, Tihaa I, Maybeck V, Balfanz S, Baumann A. Kempmann A, et al. Int J Mol Sci. 2022 Jun 13;23(12):6593. doi: 10.3390/ijms23126593. Int J Mol Sci. 2022. PMID: 35743038 Free PMC article. - Targeting the retinoblastoma protein by MC007L, gene product of the molluscum contagiosum virus: detection of a novel virus-cell interaction by a member of the poxviruses.
Mohr S, Grandemange S, Massimi P, Darai G, Banks L, Martinou JC, Zeier M, Muranyi W. Mohr S, et al. J Virol. 2008 Nov;82(21):10625-33. doi: 10.1128/JVI.01187-08. Epub 2008 Aug 13. J Virol. 2008. PMID: 18701596 Free PMC article.
References
- Anandatheerthavarada H.K., Biswas G., Mullick J., Sepuri N.B.V., Otvos L., Pain D., Avadhani N.G. Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP–dependent phosphorylation as Ser128. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:5494–5504. - PMC - PubMed
- Armstrong L.C., Komiya T., Bergman B.E., Mihara K., Bornstein P. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J. Biol. Chem. 1997;272:6510–6518. - PubMed
- Cao W., Douglas M.G. Biogenesis of ISP6, a small carboxyterminal anchored protein of the receptor complex of the mitochondrial outer membrane. J. Biol. Chem. 1995;270:5674–5679. - PubMed
- Dietmeier K., Hönlinger A., Bömer U., Dekker P.J.T., Eckerskorn C., Lottspeich F., Kübrich M., Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases