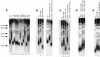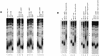Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment - PubMed (original) (raw)
Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment
N Grandin et al. Mol Cell Biol. 2000 Nov.
Abstract
The Saccharomyces cerevisiae CDC13 protein binds single-strand telomeric DNA. Here we report the isolation of new mutant alleles of CDC13 that confer either abnormal telomere lengthening or telomere shortening. This deregulation not only depended on telomerase (Est2/TLC1) and Est1, a direct regulator of telomerase, but also on the yeast Ku proteins, yKu70/Hdf1 and yKu80/Hdf2, that have been previously implicated in DNA repair and telomere maintenance. Expression of a Cdc13-yKu70 fusion protein resulted in telomere elongation, similar to that produced by a Cdc13-Est1 fusion, thus suggesting that yKu70 might promote Cdc13-mediated telomerase recruitment. We also demonstrate that Stn1 is an inhibitor of telomerase recruitment by Cdc13, based both on STN1 overexpression and Cdc13-Stn1 fusion experiments. We propose that accurate regulation of telomerase recruitment by Cdc13 results from a coordinated balance between positive control by yKu70 and negative control by Stn1. Our results represent the first evidence of a direct control of the telomerase-loading function of Cdc13 by a double-strand telomeric DNA-binding complex.
Figures
FIG. 1
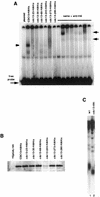
Telomere lengthening associated with the cdc13 alleles relies on telomerase activity and requires Est1. (A) Telomere Southern blot analysis of novel non-temperature-sensitive cdc13 mutant alleles generated by PCR mutagenesis (see Materials and Methods) reveals the possibility of a large panel of telomere size deregulation when CDC13 is mutated (lanes 2 to 14) compared to a wild-type strain (lane 1). Here, the cdc13 alleles (names of which are indicated on top of the figure) were present on a single-copy centromeric plasmid in a cdc13-1 mutant. These mutant strains were grown at 32°C to inactivate Cdc13-1. (B) Augmentation of telomerase activity is responsible for telomere elongation in the new cdc13 mutant strains described here. TLC1, the RNA subunit of telomerase, was disrupted directly in a cdc13-109i mutant or in a cdc13-1 mutant harboring YCp-cdc13-276. The resulting strains no longer displayed the telomere elongation conferred by the cdc13-109 and cdc13-276 mutations, as measured here after approximately 50 generations (compare lane 2 to lane 3 and lane 5 to lane 6). Telomere lengths in wild-type strains (lanes 1 and 4) are shown for comparison. (C) Telomere elongation in the cdc13 mutants necessitates the presence of Est1, a regulator of telomerase. The EST1 gene was genetically disrupted in cdc13-109i or in cdc13-1 YCp-cdc13-276 and telomere size measured after approximately 50 generations. Absence of telomere lengthening in the resulting double mutants (compare lane 2 to lane 3 and lane 4 to lane 5) was observed. (D) The Cdc13-109 and Cdc13-231 mutant proteins can sustain growth on their own when expressed from a single-copy plasmid in a strain disrupted for CDC13 (cdc13::TRP1). Both Cdc13-109 (lane 3) and Cdc13-231 (lane 4) conferred telomere lengthening compared to a wild-type strain (lane 1) or a cdc13 disruptant expressing wild-type CDC13 (lane 2). (E) Expression of the temperature-sensitive cdc13-1 allele from an episomal plasmid, at the restrictive temperature of 34°C, had no incidence on the _cdc13-109_-associated telomere lengthening (compare lanes 2 and 3).
FIG. 2
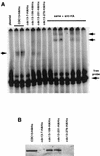
The telomere-elongating Cdc13 mutant proteins described in this study are still able to bind telomeric sequences. (A) The in vitro binding of wild-type Cdc13 and of telomere-elongating Cdc13 mutant proteins to specific TG1–3 telomeric DNA sequences was measured by assessing gel retardation of Cdc13-2HA-6His proteins (see Materials and Methods). The same reaction mixtures were incubated in parallel with a monoclonal anti-HA antibody, which generated a supershift of the complex attesting of the specificity of the DNA-protein binding reaction. Binding of the Cdc13-109–2HA–6His and Cdc13-231–2HA–6His proteins was somewhat weaker than that of wild-type Cdc13-2HA-6His, while Cdc13-7–2HA–6His and Cdc13-276–2HA–6His did not bind telomeric DNA. Unlabeled arrows indicate the position of the DNA-protein and DNA-protein-antibody complexes. (B) Immunoblot analysis, using a monoclonal anti-HA antibody, of crude extracts from cells expressing Cdc13-2HA-6His proteins under the control of the inducible GAL1 promoter was performed essentially to assess the presence of the Cdc13 mutant proteins used in band shift experiments. This analysis revealed that, in fact, Cdc13-7 and Cdc13-276 were not produced as entire proteins. Sequence analysis of these cdc13 alleles revealed the presence of a stop codon, generated during mutagenesis, upstream of the natural stop codon (see text).
FIG. 3
Mutations in YKU70 or YKU80 have a larger suppressing effect on _cdc13-109_- or _cdc13-276_-associated telomere elongation than mutations in RAD50 or TEL1. (A) cdc13-109i tel1 (lane 1), cdc13-109i yku70 (lane 7), cdc13-109i rad50 (lane 10), and cdc13-109i yku80 (lane 14) double mutants were grown for approximately 100 generations, and their telomere lengths were compared to those in the corresponding single mutants (neighboring lanes in each panel for each of the four mutations) and in wild-type cells (lanes 2, 5, 9, and 13). See the text for interpretation of the data. (B) _cdc13-1 rad50_Δ, _cdc13-1 tel1_Δ, _cdc13-1 yku70_Δ, and _cdc13-1 yku80_Δ double mutants were propagated at 25°C and transformed with a single-copy plasmid harboring the cdc13-276 allele. The resulting triple mutants were grown at 34°C to inactivate Cdc13-1, and the sizes of their telomeres were measured after approximately 100 generations (lanes 3, 6, 10, and 14) and compared to those in cdc13-1 rad50, cdc13-1 tel1, cdc13-1 yku70, or cdc13-1 yku80 (lanes 2, 5, 8, and 13), respectively, at 34°C or to that in wild-type isogenic strains (lanes 1, 4, 7, and 11). Telomere lengths in cdc13-1 cells grown at 25°C (lane 12) and in cdc13-1 cells harboring cdc13-276 on a single-copy plasmid, grown at 34°C (lane 9), served as negative and positive controls, respectively. Note that _cdc13-1 yku70_Δ YCp-cdc13-276 (lane 10) and _cdc13-1 yku80_Δ YCp-cdc13-276 cells (lane 14) no longer displayed the long telomere phenotype characteristic of the cdc13-276 allele (lane 9), contrary to cdc13-1 rad50 YCp-cdc13-276 (lane 3) and cdc13-1 tel1 YCp-cdc13-276 (lane 6) which did. This interpretation is confirmed by visualizing the dots in lane 10, which highlight two bands corresponding to two non Y′ telomeres. These were also present in wild-type cells (lane 7) and _yku70_Δ cells (lane 8) but were absent from cells harboring the cdc13-276 mutation alone (lane 9). This confirmed that the telomere structure in the cdc13-1 YCp-_cdc13-276 yku70_Δ mutant no longer resembled that in cdc13-1 YCp-cdc13-276 but was instead similar to that in wild-type cells. The same held true for _yku80_Δ (lane 14) but not for _rad50_Δ (lane 3) and _tel1_Δ (lane 6) cells. See the text for further explanations. Conditions of strain culturing were as described above.
FIG. 4
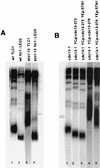
A Cdc13-yKu70 fusion protein produces telomere elongation that is dependent on Est1. (A) Expression of CDC13-EST1 or CDC13-YKU70 hybrid gene from a single-copy centromeric plasmid under the control of the CDC13 promoter for approximately 100 generations, either in a cdc13-1 strain grown at 34°C to inactivate Cdc13-1 (left panel), in a wild-type background (middle left panel), or in a _cdc13-1_Δ background (middle right panel) provoked dramatic telomere elongation (lanes 2, 5, 8, 9, 11, and 12) compared to controls (cdc13-1 cells grown at 25°C, lane 1; wild-type cells, lanes 7 and 10). A fusion consisting of the telomere-shortening Cdc13-69 mutant protein and of either wild-type Est1 or wild-type yKu70 also produced telomere elongation in _cdc13_Δ (lanes 16 and 17) or cdc13-1 at 34°C (lanes 3 and 6), therefore indicating rescue of the short telomere phenotype conferred by Cdc13-69 (lane 15). A Cdc13-Stn1 fusion (lane 13) served as a negative control. (B) Telomere elongation provoked by the Cdc13-yKu70 fusion no longer took place in the absence of Est1 (lane 6; compare to the _est1_Δ [lane 5] and wild-type [lane 4] strains). Lanes 1 and 3, in which CDC13 or EST1 alone were expressed, provided additional controls for these protein fusion experiments. See the text for further explanations. (C) Telomere elongation provoked by the Cdc13-yKu70 no longer took place in _yku80_Δ _cdc13-1_Δ cells (lane 1; compare to the YKU80+ cdc13Δ cells expressing the same fusion (lane 2) and to the wild-type [lane 3] and _yku80_Δ [lane 4] strains). This effect was observed both in a _cdc13_Δ background (lane 1) and a CDC13+ background (lane 5).
FIG. 5
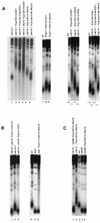
In vitro binding of telomere-shortening Cdc13 mutant proteins to specific TG1–3 telomeric DNA sequences. (A) Band shift experiments, performed as described in the legend to Fig. 2, revealed that all six Cdc13 mutant proteins were severely defective in DNA binding, with Cdc13-23–2HA-6His, Cdc13-30–2HA-6His, and Cdc13-280–2HA-6His being more affected than Cdc13-69–2HA-6His, Cdc13-243–2HA-6His, and Cdc13-273–2HA-6His, which still showed some binding. The specificity of the DNA-protein interaction was attested to by visualizing a supershift upon addition of a monoclonal anti-HA antibody to the reaction mixture. Unlabeled arrows indicate the position of the DNA-protein and DNA-protein-antibody complexes. (B) Western blot analysis of crude extracts from cells expressing Cdc13–2HA-6His proteins under the control of the inducible GAL1 promoter, using a monoclonal anti-HA antibody, revealed that basically all of the Cdc13 mutant proteins were produced to the same extent within the cell, with the exception of Cdc13-243–2HA-6His, whose levels were lower than those of the other mutant proteins. (C) The cdc13-69i strain, harboring the telomere-shortening cdc13-69 allele integrated at the CDC13 locus, exhibited telomeres shortened to the same extent as those in a cdc13-1 YCp-cdc13-69 strain grown at 34°C (Fig. 1A, lane 12) or in a _cdc13_Δ YCp-cdc13-69 strain (Fig. 4A, lane 15).
FIG. 6
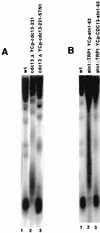
Stn1 behaves as an inhibitor of telomerase recruitment. (A) A “young” stn1-13 mutant strain (in which the stn1-13 mutation had been introduced at the STN1 locus only for a week so as to obtain cells with telomeres of nearly wild-type size) was disrupted for TLC1 and grown at 30°C for approximately 50 generations. In this stn1-13 tlc1::LEU2 strain, telomeres remained short (lane 4), of the same size as in a wild-type strain disrupted for TLC1 (lane 2), and shorter than in wild-type TLC1+ cells (lane 1), in contrast with telomeres in stn1-13 TLC1+ cells which during the same period of growth had become very long (lane 3). (B) Overexpression of STN1 diminishes the _cdc13-276_-associated telomere elongation and aggravates the _cdc13-273_-associated telomere shortening. A cdc13-1 YCp-cdc13-273 mutant was transformed with YEp-STN1, a multicopy plasmid overexpressing STN1 under the control of its natural promoter (lane 3), or with vector alone (lane 2). Both strains were grown at 32°C (to inactivate Cdc13-1), and the size of their telomeres were measured after approximately 100 generations. Short telomeres conferred by the Cdc13-273 protein (lane 2; compare to the telomere size in a cdc13-1 strain grown at 25°C [lane 1]) were further shortened following STN1 overexpression (lane 3). In addition, abnormal telomere lengthening conferred by the Cdc13-276 mutant protein (lane 4) was partially relieved when STN1 was overexpressed (lane 5).
FIG. 7
Expression of Stn1-Cdc13 fusion proteins confirm that Stn1 negatively regulates Cdc13. (A) Expression of a cdc13-231–STN1 hybrid gene from a single-copy centromeric plasmid under the control of the CDC13 promoter in a _cdc13_Δ strain grown for approximately 100 generations totally suppressed the long telomere phenotype conferred by Cdc13-231 (compare lanes 2 and 3). (B) Expression of a CDC13–stn1-63 hybrid gene in an _stn1_Δ strain under the same conditions as those described above totally suppressed the telomere lengthening phenotype conferred by Stn1-63 (compare lanes 2 and 3), which resulted in the acquisition of telomeres of wild-type size (lane 1).
FIG. 8
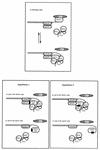
(Top) A speculative model for yKu70 and Stn1 as positive and negative regulators, respectively, of Cdc13-Est1-mediated telomerase loading in wild-type cells. Stn1, which physically associates with Cdc13, a single-strand telomeric DNA-binding protein, acts as an inhibitor of Cdc13-mediated telomerase recruitment (telomerase contains the catalytic subunit, Est2, and TLC1, the RNA template; Est1, a regulator of telomerase which binds single-stranded telomeric DNA, also physically associates with TLC1). Stn1 release from the Cdc13-Stn1 complex might represent the signal allowing interactions between Cdc13 and Est1-telomerase. According to this model, interactions between Cdc13 and the DNA repair yKu proteins, yKu70 and yKu80 (yKu70 physically associates with yKu80, which itself binds double-strand telomeric DNA), might promote efficient association of Est1-telomerase with the telomere end, thus allowing telomeric DNA addition. Re-binding of Stn1 to Cdc13 might compete with Cdc13-yKu70 or -yKu80 and Cdc13-Est1 interactions, therefore breaking interactions between Est1-telomerase and telomeric DNA and terminating the telomere replication process. (Bottom) Two hypotheses are proposed to explain the deregulation of telomere length conferred by the telomere-elongating cdc13 alleles described in the present study (cdc13-109, which has been used in most of the experiments presented here, has been chosen for illustration). For both hypotheses, the situation has been envisioned either in the presence of the cdc13-109i mutation alone (cdc13-109 YKU70 cells) or in the simultaneous presence of the cdc13-109i and the _yku70_Δ mutations (cdc13-109 yku70 cells). Full ovals represent a higher than normal physical association between the deregulated Cdc13-109 mutant protein and yKu70 (hypothesis 1, left) or between Cdc13-109 and Est1 (hypothesis 2, right). In all of the configurations represented here, Stn1 is in the off position, physically apart from Cdc13, the position that presumably allows telomerase recruitment by Cdc13. In hypothesis 1 (left), constitutive interactions between Cdc13 and either yKu70 or yKu80 provokes recruitment of telomerase at higher than normal levels, thus leading to telomere lengthening (top), while the absence of yKu70 presumably results in inefficient recruitment of Est1-telomerase, thus suppressing _cdc13-109_-induced telomere lengthening (bottom). In hypothesis 2 (right), constitutive interactions between Cdc13 and Est1 also provoke recruitment of telomerase at higher-than-normal levels and leads to telomere lengthening (top), but this time the absence of yKu70 presumably generates an abnormal single-stranded telomeric DNA extension which competes for Cdc13-109–Est1 interactions and results in inefficient recruitment of Est1-telomerase and suppression of the _cdc13-109_-induced telomere lengthening (bottom). See the text for further explanations.
Similar articles
- Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13.
Grandin N, Damon C, Charbonneau M. Grandin N, et al. EMBO J. 2001 Mar 1;20(5):1173-83. doi: 10.1093/emboj/20.5.1173. EMBO J. 2001. PMID: 11230140 Free PMC article. - Est1 and Cdc13 as comediators of telomerase access.
Evans SK, Lundblad V. Evans SK, et al. Science. 1999 Oct 1;286(5437):117-20. doi: 10.1126/science.286.5437.117. Science. 1999. PMID: 10506558 - Telomere maintenance is dependent on activities required for end repair of double-strand breaks.
Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Nugent CI, et al. Curr Biol. 1998 May 21;8(11):657-60. doi: 10.1016/s0960-9822(98)70253-2. Curr Biol. 1998. PMID: 9635193 - Telomeres--unsticky ends.
Shore D. Shore D. Science. 1998 Sep 18;281(5384):1818-9. doi: 10.1126/science.281.5384.1818. Science. 1998. PMID: 9776685 Review. No abstract available. - Regulation of telomerase by telomeric proteins.
Smogorzewska A, de Lange T. Smogorzewska A, et al. Annu Rev Biochem. 2004;73:177-208. doi: 10.1146/annurev.biochem.73.071403.160049. Annu Rev Biochem. 2004. PMID: 15189140 Review.
Cited by
- Dysfunction of Telomeric Cdc13-Stn1-Ten1 Simultaneously Activates DNA Damage and Spindle Checkpoints.
Grandin N, Charbonneau M. Grandin N, et al. Cells. 2024 Sep 25;13(19):1605. doi: 10.3390/cells13191605. Cells. 2024. PMID: 39404369 Free PMC article. - DNA-dependent protein kinase in telomere maintenance and protection.
Sui J, Zhang S, Chen BPC. Sui J, et al. Cell Mol Biol Lett. 2020 Jan 17;25:2. doi: 10.1186/s11658-020-0199-0. eCollection 2020. Cell Mol Biol Lett. 2020. PMID: 31988640 Free PMC article. Review. - The telomeric Cdc13-Stn1-Ten1 complex regulates RNA polymerase II transcription.
Calvo O, Grandin N, Jordán-Pla A, Miñambres E, González-Polo N, Pérez-Ortín JE, Charbonneau M. Calvo O, et al. Nucleic Acids Res. 2019 Jul 9;47(12):6250-6268. doi: 10.1093/nar/gkz279. Nucleic Acids Res. 2019. PMID: 31006804 Free PMC article. - Vps74 Connects the Golgi Apparatus and Telomeres in Saccharomyces cerevisiae.
Rodrigues J, Banks P, Lydall D. Rodrigues J, et al. G3 (Bethesda). 2018 May 4;8(5):1807-1816. doi: 10.1534/g3.118.200172. G3 (Bethesda). 2018. PMID: 29593073 Free PMC article. - The evolutionarily conserved factor Sus1/ENY2 plays a role in telomere length maintenance.
Galán A, García-Oliver E, Nuño-Cabanes C, Rubinstein L, Kupiec M, Rodríguez-Navarro S. Galán A, et al. Curr Genet. 2018 Jun;64(3):635-644. doi: 10.1007/s00294-017-0778-4. Epub 2017 Nov 7. Curr Genet. 2018. PMID: 29116388
References
- Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1998.
- Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous