Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha - PubMed (original) (raw)
Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha
J Solsbacher et al. Mol Cell Biol. 2000 Nov.
Abstract
Import of proteins containing a classical nuclear localization signal (NLS) into the nucleus is mediated by importin alpha and importin beta. Srp1p, the Saccharomyces cerevisiae homologue of importin alpha, returns from the nucleus in a complex with its export factor Cse1p and with Gsp1p (yeast Ran) in its GTP-bound state. We studied the role of the nucleoporin Nup2p in the transport cycle of Srp1p. Cells lacking NUP2 show a specific defect in both NLS import and Srp1p export, indicating that Nup2p is required for efficient bidirectional transport of Srp1p across the nuclear pore complex (NPC). Nup2p is located at the nuclear side of the central gated channel of the NPC and provides a binding site for Srp1p via its amino-terminal domain. We show that Nup2p effectively releases the NLS protein from importin alpha-importin and beta and strongly binds to the importin heterodimer via Srp1p. Kap95p (importin beta) is released from this complex by a direct interaction with Gsp1p-GTP. These data suggest that besides Gsp1p, which disassembles the NLS-importin alpha-importin beta complex upon binding to Kap95p in the nucleus, Nup2p can also dissociate the import complex by binding to Srp1p. We also show data indicating that Nup1p, a relative of Nup2p, plays a similar role in termination of NLS import. Cse1p and Gsp1p-GTP release Srp1p from Nup2p, which suggests that the Srp1p export complex can be formed directly at the NPC. The changed distribution of Cse1p at the NPC in nup2 mutants also supports a role for Nup2p in Srp1p export from the nucleus.
Figures
FIG. 1
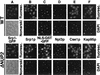
Export of Srp1p and Cse1p as well as NLS protein import are defective in cells lacking NUP2. (A) Yeast cells carrying the SRP1-GFP allele and a deletion of NUP2 (Δ_NUP2_ strain GSY618) were transformed with plasmid pGS248 encoding Nup2p (top) or with the CEN LEU2 vector pRS315 (bottom). Cells were grown in liquid medium at 30°C. Srp1-GFP was visualized by fluorescence microscopy (fluoresc.) and also viewed by Nomarski optics. (B to F) Wild-type (WT; W303), Δ_NUP2_ (JLY506) cells or the same cells transformed with the reporter plasmid pGS422, encoding SV40 NLS-GST-GFP (C), were grown at 30°C. Synthesis of NLS-GST-GFP was induced by 2% galactose for 3 h (C). Cells were prepared for immunofluorescence microscopy and incubated with antibodies against Srp1p (B), GFP (C), Npl3p (D), Cse1p (E), and Kap95p (F). Antibodies were detected by Texas red-conjugated IgG, and DNA was stained with DAPI.
FIG. 2
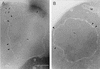
Srp1p fails to localize to the nuclear side of the NPC in cells lacking NUP2. Wild-type (W303; A) and Δ_NUP2_ (JLY506; B) cells were grown in liquid medium at 30°C and prepared for immunoelectron microscopy. Ultrathin cryosections of whole cells were incubated with Srp1p-specific antibodies and with secondary antibody-gold complexes (10 nm; Dianova). Intracellular structures: nucleus (N), nuclear pores (arrowheads), mitochondrion (M), endoplasmic reticulum (ER), and vacuole (V). Bars represent 500 nm.
FIG. 3
Nup2p and Nup1p localize to the nuclear side of the NPC. The genomic copies of NUP2 and NUP1 were replaced by NUP2-GFP (GSY627; A) and NUP1-GFP (GSY652; B), respectively. Cells were grown in liquid medium at 30°C and prepared for immunoelectron microscopy. Ultrathin cryosections were incubated with polyclonal anti-GFP antibodies (Clontech) and with gold-labeled secondary antibodies. Intracellular structures: nucleus (N, n), cytoplasm (c), nuclear pores (Np), mitochondrion (M), and endoplasmic reticulum (ER). The bar represents 100 nm.
FIG. 4
NLS protein import is defective in cells deleted for NUP1. Wild-type (W303; A) and Δ_NUP1_ (LDY461 [23]; B to D) cells were transformed with the reporter plasmid pGS420, encoding NLS-GST-GFP (A to C), or pGS846, encoding GST-GFP-MATα2p (D). Cells were grown in liquid medium containing 2% raffinose and incubated with 2% galactose for 3 h at 25°C (B) or incubated with 2% galactose for 90 min at 25°C and shifted to 37°C for a further 90 min (A, C, and D). GFP fluorescence was visualized microscopically.
FIG. 5
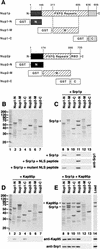
Srp1p and Kap95p directly interact with Nup1p and Nup2p. (A) Domain structures of Nup1p and Nup2p. Nup1p and Nup2p are related in their central domains that contain the FXFG repeats. Nup2p is the only yeast nucleoporin possessing a Ran-binding domain (RBD) of the Yrb1p/RanBP1 type. The residues fused to GST are indicated. The Nup1-M construct contains repeats 6 to 22 of the 29 Nup1p FXFG motifs. Nup2-M contains all 15 Nup2p FXFG repeats. (B to E) Recombinant fusion proteins of GST and the indicated fragments of Nup1p or Nup2p (4 μg) were immobilized to glutathione-Sepharose and incubated with buffer alone (B), with 5 μg of purified Srp1p (C), with 7 μg of purified Kap95p (D), or with Srp1p and Kap95p (E) for 30 min at 4°C. After three washes, bound material was eluted with SDS sample buffer and analyzed by SDS-PAGE and Coomassie blue staining. Positions of molecular weight markers are shown in kilodaltons (lane 1). The eluates as well as Srp1p and Kap95p, representing 25% of the load (lane 14), were also analyzed by immunoblotting with antibodies against Srp1p and Kap95p. GST-Nup fusion proteins were incubated with Srp1p and a 1,000-fold molar excess of SV40 large T-antigen NLS peptides (CTPPKKKRKV) or mutant NLS peptides (CTPPKTKRKV). Bound material was analyzed by immunoblotting with Srp1p-specific antibodies (C).
FIG. 6
Nup2p displaces the NLS protein from the NLS-Srp1p-Kap95p complex. (A) A fusion of GST to Prp20p (8 μg) was immobilized to glutathione-Sepharose and incubated with 8 μg of Srp1p and 12 μg of Kap95p for 30 min at 4°C. After three washes, bound material was further incubated with buffer alone (lanes 7 and 12), 8 μg of Nup2-N (lanes 8 and 13), SV40 NLS peptide (1,000-fold molar excess) (lanes 9 and 13), 10 μg of Gsp1p-GTP (lanes 10 and 15), or Gsp1p-GDP (lane 11). After incubation at 4°C for 10 min, bound (lanes 7 to 11) and unbound (lanes 12 to 15) material was separated and analyzed by SDS-PAGE and Coomassie blue staining. Molecular weight markers (positions indicated in kilodaltons) and loads of recombinant proteins are shown in lanes 1 to 6. (B) GST–Nup2-N (10 μg) (lanes 1 to 5) or GST-Srp1p (8 μg) (lanes 7 to 15) was immobilized to glutathione-Sepharose and incubated for 30 min at 4°C with 8 μg of Srp1p, 12 μg of Kap95p, 10 μg of Prp20p, or 8 μg of Nup2-N as indicated. After three washes, bound material was analyzed by SDS-PAGE and Coomassie blue staining. Samples containing the Prp20p-Srp1p-Kap95p complex were further incubated with buffer (lane 13), with Nup2-N (lane 14), or with SV40 NLS peptide (1,000-fold molar excess) (lane 15). After incubation at 4°C for 10 min, bound material was examined by SDS-PAGE and Coomassie blue staining. Prp20p purified via an N-terminal His6 tag (50% input) was loaded in lane 6. All samples were also analyzed by immunoblotting with anti-Prp20p antibodies.
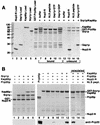
FIG. 7
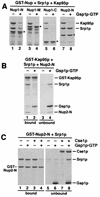
Kap95p complexed to Gsp1p-GTP does not bind to Nup1/2p-associated Srp1p. (A) Fusions of GST to Nup1-N, Nup1-M, Nup1-C, and Nup2-N (asterisks) were incubated with Srp1p and Kap95p as described for Fig. 5E. After three washes, buffer (lanes 1, 3, 5, and 7) or 10 μg of Gsp1p-GTP (lanes 2, 4, 6, and 8) was added, and the reaction mixtures were further incubated for 15 min at 25°C. After washing, bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE and Coomassie blue staining. (B) Immobilized GST-Kap95p (6 μg) was incubated with 6 μg of Srp1p and 4 μg of Nup2-N for 30 min at 4°C. After three washes, the Kap95p–Srp1p–Nup2-N complex was further incubated with buffer alone (lanes 1 and 3) or with 10 μg of Gsp1p-GTP (lanes 2 and 4) for 15 min. After washing, bound (lanes 1 and 2) and unbound (lanes 3 and 4) material was analyzed by SDS-PAGE and Coomassie blue staining. (C) Cse1p and Gsp1p-GTP release Srp1p from Nup2-N. Immobilized GST–Nup2-N (4 μg per reaction) preincubated with 8 μg of Srp1p was incubated with buffer alone (lanes 1 and 5), with 10 μg of Gsp1p-GTP (lanes 2 and 6), with 12 μg of Cse1p (lanes 3 and 7), or with Gsp1p-GTP and Cse1p (lanes 4 and 8) for 15 min at 25°C. Proteins bound to glutathione-Sepharose (lanes 1 to 4) and the unbound material (lanes 5 to 8) was analyzed by SDS-PAGE and Coomassie blue staining.
FIG. 8
Localization of Cse1p at the NPC in wild-type and Δ_NUP2_ cells. Wild-type cells (GSY580; A) and Δ_NUP2_ cells (GSY777; B) carrying genomic copies of GFP-CSE1 were grown in liquid medium at 30°C and prepared for immunoelectron microscopy. Ultrathin cryosections of whole cells were incubated with anti-GFP antibodies and with secondary antibodies conjugated to 10-nm gold particles. Intracellular structures: nucleus (n), cytoplasm (c), and nuclear pores (arrows). Bars represent 100 nm.
FIG. 9
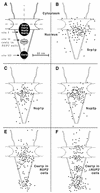
(A) Schematic representation of the NPC with the major localization peaks of Srp1p, Nup1p, Nup2p, and Cse1p. All of these proteins colocalize at the nuclear side of the central gated channel in wild-type cells (site I). Cse1p is also present at the distal ring of the nuclear basket (site III). A third Cse1p peak in the central region of the basket (site II) appears only in Δ_NUP2_ cells. The dimensions of the NPC are taken from references and . The vertical and horizontal axes are indicated by dashed lines. (B to F) Gold particles corresponding to Srp1p, Nup1p, Nup2p, and Cse1p labeling were projected onto the NPC model. (G to L) Quantification of the distribution at the NPC of gold particles corresponding to Srp1p (128 gold particles), Nup1-GFP (89 gold particles), Nup2-GFP (110 gold particles), GFP-Cse1p in wild-type cells (135 gold particles), and GFP-Cse1p in Δ_NUP2_ cells (180 gold particles).
FIG. 9
(A) Schematic representation of the NPC with the major localization peaks of Srp1p, Nup1p, Nup2p, and Cse1p. All of these proteins colocalize at the nuclear side of the central gated channel in wild-type cells (site I). Cse1p is also present at the distal ring of the nuclear basket (site III). A third Cse1p peak in the central region of the basket (site II) appears only in Δ_NUP2_ cells. The dimensions of the NPC are taken from references and . The vertical and horizontal axes are indicated by dashed lines. (B to F) Gold particles corresponding to Srp1p, Nup1p, Nup2p, and Cse1p labeling were projected onto the NPC model. (G to L) Quantification of the distribution at the NPC of gold particles corresponding to Srp1p (128 gold particles), Nup1-GFP (89 gold particles), Nup2-GFP (110 gold particles), GFP-Cse1p in wild-type cells (135 gold particles), and GFP-Cse1p in Δ_NUP2_ cells (180 gold particles).
Similar articles
- The yeast nucleoporin Nup2p is involved in nuclear export of importin alpha/Srp1p.
Booth JW, Belanger KD, Sannella MI, Davis LI. Booth JW, et al. J Biol Chem. 1999 Nov 5;274(45):32360-7. doi: 10.1074/jbc.274.45.32360. J Biol Chem. 1999. PMID: 10542277 - Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-alpha export.
Hood JK, Casolari JM, Silver PA. Hood JK, et al. J Cell Sci. 2000 Apr;113 ( Pt 8):1471-80. doi: 10.1242/jcs.113.8.1471. J Cell Sci. 2000. PMID: 10725229 - The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex.
Denning D, Mykytka B, Allen NP, Huang L, Al Burlingame, Rexach M. Denning D, et al. J Cell Biol. 2001 Sep 3;154(5):937-50. doi: 10.1083/jcb.200101007. J Cell Biol. 2001. PMID: 11535617 Free PMC article. - Molecular mechanisms of nuclear protein transport.
Moroianu J. Moroianu J. Crit Rev Eukaryot Gene Expr. 1997;7(1-2):61-72. doi: 10.1615/critreveukargeneexpr.v7.i1-2.40. Crit Rev Eukaryot Gene Expr. 1997. PMID: 9034715 Review. - The importin β binding domain as a master regulator of nucleocytoplasmic transport.
Lott K, Cingolani G. Lott K, et al. Biochim Biophys Acta. 2011 Sep;1813(9):1578-92. doi: 10.1016/j.bbamcr.2010.10.012. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029753 Free PMC article. Review.
Cited by
- Recent Advances on the Structure and Function of RNA Acetyltransferase Kre33/NAT10.
Sleiman S, Dragon F. Sleiman S, et al. Cells. 2019 Sep 5;8(9):1035. doi: 10.3390/cells8091035. Cells. 2019. PMID: 31491951 Free PMC article. Review. - Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex.
Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Dilworth DJ, et al. J Cell Biol. 2001 Jun 25;153(7):1465-78. doi: 10.1083/jcb.153.7.1465. J Cell Biol. 2001. PMID: 11425876 Free PMC article. - Sumoylation regulates Kap114-mediated nuclear transport.
Rothenbusch U, Sawatzki M, Chang Y, Caesar S, Schlenstedt G. Rothenbusch U, et al. EMBO J. 2012 May 30;31(11):2461-72. doi: 10.1038/emboj.2012.102. Epub 2012 May 4. EMBO J. 2012. PMID: 22562154 Free PMC article. - A nuclear pore sub-complex restricts the propagation of Ty retrotransposons by limiting their transcription.
Bonnet A, Chaput C, Palmic N, Palancade B, Lesage P. Bonnet A, et al. PLoS Genet. 2021 Nov 1;17(11):e1009889. doi: 10.1371/journal.pgen.1009889. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34723966 Free PMC article. - Nup2 performs diverse interphase functions in Aspergillus nidulans.
Suresh S, Markossian S, Osmani AH, Osmani SA. Suresh S, et al. Mol Biol Cell. 2018 Dec 15;29(26):3144-3154. doi: 10.1091/mbc.E18-04-0223. Epub 2018 Oct 24. Mol Biol Cell. 2018. PMID: 30355026 Free PMC article.
References
- Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-β. Cell. 2000;102:99–108. - PubMed
- Booth J W, Belanger K D, Sannella M I, Davis L I. The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J Biol Chem. 1999;274:32360–32367. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous