The circadian clock mutation alters sleep homeostasis in the mouse - PubMed (original) (raw)
The circadian clock mutation alters sleep homeostasis in the mouse
E Naylor et al. J Neurosci. 2000.
Abstract
The onset and duration of sleep are thought to be primarily under the control of a homeostatic mechanism affected by previous periods of wake and sleep and a circadian timing mechanism that partitions wake and sleep into different portions of the day and night. The mouse Clock mutation induces pronounced changes in overall circadian organization. We sought to determine whether this genetic disruption of circadian timing would affect sleep homeostasis. The Clock mutation affected a number of sleep parameters during entrainment to a 12 hr light/dark (LD 12:12) cycle, when animals were free-running in constant darkness (DD), and during recovery from 6 hr of sleep deprivation in LD 12:12. In particular, in LD 12:12, heterozygous and homozygous Clock mutants slept, respectively, approximately 1 and approximately 2 hr less than wild-type mice, and they had 25 and 51% smaller increases in rapid eye movement (REM) sleep during 24 hr recovery, respectively, than wild-type mice. The effects of the mutation on sleep are not readily attributable to differential entrainment to LD 12:12 because the baseline sleep differences between genotypes were also present when animals were free-running in DD. These results indicate that genetic alterations of the circadian clock system and/or its regulatory genes are likely to have widespread effects on a variety of sleep and wake parameters, including the homeostatic regulation of sleep.
Figures
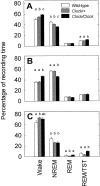
Fig. 1.
Percentage of recording time spent in the sleep states for all three genotypes. The four bar groups represent wake, NREM or REM sleep, and the percentage of sleep time spent in REM sleep: a measure of the REM/NREM ratio. A shows sleep during the entire 24 hr LD baseline period, whereas the other graphs further break this baseline period into the 12 hr light period (B) and the 12 hr dark period (C). a–c indicate significant pairwise differences between groups (p < 0.05, Tukey–Kramer post hoc tests).
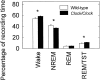
Fig. 2.
Percentage of circadian period spent in various sleep states for wild-type and homozygous Clock mice housed in constant darkness. TST, Total sleep time.
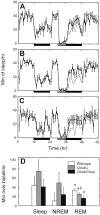
Fig. 3.
Response to sleep deprivation. Minutes of sleep per hour during baseline, sleep deprivation (gray bar), and recovery periods for wild-type (A), Clock heterozygous (B), and Clock homozygous (C) mice. Black bars represent times of lights off. For comparison, the baseline sleep amounts have been double-plotted in gray during the recovery period.D, The number of additional minutes of sleep over each animal's equivalent baseline for the entire 24 hr LD recovery period.a and b indicate significant pairwise differences between groups (p < 0.05, Tukey–Kramer post hoc tests).
Similar articles
- Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation.
Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Laposky A, et al. Sleep. 2005 Apr;28(4):395-409. doi: 10.1093/sleep/28.4.395. Sleep. 2005. PMID: 16171284 - Altered circadian and homeostatic sleep regulation in prokineticin 2-deficient mice.
Hu WP, Li JD, Zhang C, Boehmer L, Siegel JM, Zhou QY. Hu WP, et al. Sleep. 2007 Mar;30(3):247-56. Sleep. 2007. PMID: 17425220 Free PMC article. - The circadian clock gene Csnk1e regulates rapid eye movement sleep amount, and nonrapid eye movement sleep architecture in mice.
Zhou L, Bryant CD, Loudon A, Palmer AA, Vitaterna MH, Turek FW. Zhou L, et al. Sleep. 2014 Apr 1;37(4):785-93, 793A-793C. doi: 10.5665/sleep.3590. Sleep. 2014. PMID: 24744456 Free PMC article. - Circadian clock genes and sleep homeostasis.
Franken P, Dijk DJ. Franken P, et al. Eur J Neurosci. 2009 May;29(9):1820-9. doi: 10.1111/j.1460-9568.2009.06723.x. Epub 2009 Apr 28. Eur J Neurosci. 2009. PMID: 19473235 Review. - Clock-Sleep Communication.
Pandi-Perumal SR, Paul S, Saravanan KM, Namasivayam GP, Chidambaram SB. Pandi-Perumal SR, et al. Curr Mol Med. 2025;25(4):399-415. doi: 10.2174/0115665240305615240630113434. Curr Mol Med. 2025. PMID: 39694958 Review.
Cited by
- Manipulating the circadian and sleep cycles to protect against metabolic disease.
Nohara K, Yoo SH, Chen ZJ. Nohara K, et al. Front Endocrinol (Lausanne). 2015 Mar 23;6:35. doi: 10.3389/fendo.2015.00035. eCollection 2015. Front Endocrinol (Lausanne). 2015. PMID: 25852644 Free PMC article. Review. - Unraveling the Evolutionary Determinants of Sleep.
Joiner WJ. Joiner WJ. Curr Biol. 2016 Oct 24;26(20):R1073-R1087. doi: 10.1016/j.cub.2016.08.068. Curr Biol. 2016. PMID: 27780049 Free PMC article. Review. - Scale invariance in the dynamics of spontaneous behavior.
Proekt A, Banavar JR, Maritan A, Pfaff DW. Proekt A, et al. Proc Natl Acad Sci U S A. 2012 Jun 26;109(26):10564-9. doi: 10.1073/pnas.1206894109. Epub 2012 Jun 7. Proc Natl Acad Sci U S A. 2012. PMID: 22679281 Free PMC article. - Lactate as a biomarker for sleep.
Naylor E, Aillon DV, Barrett BS, Wilson GS, Johnson DA, Johnson DA, Harmon HP, Gabbert S, Petillo PA. Naylor E, et al. Sleep. 2012 Sep 1;35(9):1209-22. doi: 10.5665/sleep.2072. Sleep. 2012. PMID: 22942499 Free PMC article. - Technologies of sleep research.
Deboer T. Deboer T. Cell Mol Life Sci. 2007 May;64(10):1227-35. doi: 10.1007/s00018-007-6533-0. Cell Mol Life Sci. 2007. PMID: 17364139 Free PMC article. Review.
References
- Aeschbach D, Cajochen C, Landolt H, Borbely AA. Homeostatic sleep regulation in habitual short sleepers and long sleepers. Am J Physiol. 1996;270:R41–R53. - PubMed
- Akerstedt T, Gillberg M. The circadian variation of experimentally displaced sleep. Sleep. 1981;4:159–169. - PubMed
- Andretic R, Chaney S, Hirsh J. Requirement of circadian genes for cocaine sensitization in Drosophila. Science. 1999;285:1066–1068. - PubMed
- Benington JH, Heller HC. REM-sleep timing is controlled homeostatically by accumulation of REM- sleep propensity in non-REM sleep. Am J Physiol. 1994;266:R1992–R2000. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 AG011412/AG/NIA NIH HHS/United States
- HL-T32-07909/HL/NHLBI NIH HHS/United States
- HL/MH-RO1-59598/HL/NHLBI NIH HHS/United States
- P01-AG-11412/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials