A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions - PubMed (original) (raw)
A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions
M D Michelitsch et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Glutamine/asparagine (Q/N)-rich domains have a high propensity to form self-propagating amyloid fibrils. This phenomenon underlies both prion-based inheritance in yeast and aggregation of a number of proteins involved in human neurodegenerative diseases. To examine the prevalence of this phenomenon, complete proteomic sequences of 31 organisms and several incomplete proteomic sequences were examined for Q/N-rich regions. We found that Q/N-rich regions are essentially absent from the thermophilic bacterial and archaeal proteomes. Moreover, the average Q/N content of the proteins in these organisms is markedly lower than in mesophilic bacteria and eukaryotes. Mesophilic bacterial proteomes contain a small number (0-4) of proteins with Q/N-rich regions. Remarkably, Q/N-rich domains are found in a much larger number of eukaryotic proteins (107-472 per proteome) with diverse biochemical functions. Analyses of these regions argue they have been evolutionarily selected perhaps as modular "polar zipper" protein-protein interaction domains. These data also provide a large pool of potential novel prion-forming proteins, two of which have recently been shown to behave as prions in yeast, thus suggesting that aggregation or prion-like regulation of protein function may be a normal regulatory process for many eukaryotic proteins with a wide variety of functions.
Figures
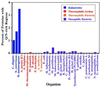
Figure 1
Many eukaryotic proteins contain Q/N-rich regions. The percent of ORFs with a region of 80 consecutive amino acids containing a Q/N content of at least 30 was determined for the indicated proteomes.
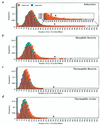
Figure 2
Expected and observed distributions of Q/N content of the best 80 mers across mesophilic and thermophilic proteomes. A comparison of the expected and observed distributions of Q/N contents across the most Q/N-rich regions of (a) eukaryotes, (b) mesophilic bacteria, (c) thermophilic bacteria, and (d) archaea. Arrows indicate the minimum Q/N content required for a domain to be considered Q/N rich. (a Inset) Magnification of the observed and expected distributions of Q/N-rich regions. Expected values were calculated on the basis of the average Q/N content per best 80 mer for each of the four types of organisms.
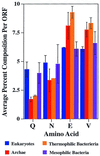
Figure 3
Thermophiles have a lower Q/N content and higher glutamate and valine content than mesophiles. The average percent amino acid content per ORF per proteome was determined for the completed proteomes. Shown are glutamine (Q), asparagine (N), glutamate (E), and valine (V). The other amino acids showed little variation. Error bars indicate the observed variance among the indicated class.
Similar articles
- Controlling the prion propensity of glutamine/asparagine-rich proteins.
Paul KR, Ross ED. Paul KR, et al. Prion. 2015;9(5):347-54. doi: 10.1080/19336896.2015.1111506. Prion. 2015. PMID: 26555096 Free PMC article. - Hyper-expansion of asparagines correlates with an abundance of proteins with prion-like domains in Plasmodium falciparum.
Singh GP, Chandra BR, Bhattacharya A, Akhouri RR, Singh SK, Sharma A. Singh GP, et al. Mol Biochem Parasitol. 2004 Oct;137(2):307-19. doi: 10.1016/j.molbiopara.2004.05.016. Mol Biochem Parasitol. 2004. PMID: 15383301 - Fitting yeast and mammalian prion aggregation kinetic data with the Finke-Watzky two-step model of nucleation and autocatalytic growth.
Watzky MA, Morris AM, Ross ED, Finke RG. Watzky MA, et al. Biochemistry. 2008 Oct 7;47(40):10790-800. doi: 10.1021/bi800726m. Epub 2008 Sep 12. Biochemistry. 2008. PMID: 18785757 - Assembly of the asparagine- and glutamine-rich yeast prions into protein fibrils.
Bousset L, Savistchenko J, Melki R. Bousset L, et al. Curr Alzheimer Res. 2008 Jun;5(3):251-9. doi: 10.2174/156720508784533303. Curr Alzheimer Res. 2008. PMID: 18537542 Review. - Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquisition?
Chernoff YO. Chernoff YO. Curr Opin Chem Biol. 2004 Dec;8(6):665-71. doi: 10.1016/j.cbpa.2004.09.002. Curr Opin Chem Biol. 2004. PMID: 15556413 Review.
Cited by
- Amyloid-mediated sequestration of essential proteins contributes to mutant huntingtin toxicity in yeast.
Kochneva-Pervukhova NV, Alexandrov AI, Ter-Avanesyan MD. Kochneva-Pervukhova NV, et al. PLoS One. 2012;7(1):e29832. doi: 10.1371/journal.pone.0029832. Epub 2012 Jan 11. PLoS One. 2012. PMID: 22253794 Free PMC article. - Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains.
Guo L, Han A, Bates DL, Cao J, Chen L. Guo L, et al. Proc Natl Acad Sci U S A. 2007 Mar 13;104(11):4297-302. doi: 10.1073/pnas.0608041104. Epub 2007 Mar 5. Proc Natl Acad Sci U S A. 2007. PMID: 17360518 Free PMC article. - A network of protein interactions determines polyglutamine toxicity.
Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. Duennwald ML, et al. Proc Natl Acad Sci U S A. 2006 Jul 18;103(29):11051-6. doi: 10.1073/pnas.0604548103. Epub 2006 Jul 10. Proc Natl Acad Sci U S A. 2006. PMID: 16832049 Free PMC article. - A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes.
Harrison PM, Gerstein M. Harrison PM, et al. Genome Biol. 2003;4(6):R40. doi: 10.1186/gb-2003-4-6-r40. Epub 2003 May 30. Genome Biol. 2003. PMID: 12801414 Free PMC article. - Psp2, a novel regulator of autophagy that promotes autophagy-related protein translation.
Yin Z, Liu X, Ariosa A, Huang H, Jin M, Karbstein K, Klionsky DJ. Yin Z, et al. Cell Res. 2019 Dec;29(12):994-1008. doi: 10.1038/s41422-019-0246-4. Epub 2019 Oct 30. Cell Res. 2019. PMID: 31666677 Free PMC article.
References
- Perutz M F. Trends Biochem Sci. 1999;24:58–63. - PubMed
- Ross C A, Margolis R L, Becher M W, Wood J D, Engelender S, Cooper J K, Sharp A H. Prog Brain Res. 1998;117:397–419. - PubMed
- DePace A H, Santoso A, Hillner P, Weissman J S. Cell. 1998;93:1241–1252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases