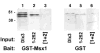The Dlx3 protein harbors basic residues required for nuclear localization, transcriptional activity and binding to Msx1 - PubMed (original) (raw)
The Dlx3 protein harbors basic residues required for nuclear localization, transcriptional activity and binding to Msx1
J T Bryan et al. J Cell Sci. 2000 Nov.
Abstract
The murine Dlx3 protein is a putative transcriptional activator that has been implicated during development and differentiation of epithelial tissue. Dlx3 contains a homeodomain and mutational analysis has revealed two regions, one N-terminal and one C-terminal to the homeodomain, that act as transcriptional activators in a yeast one-hybrid assay. In addition to transactivation, data are presented to demonstrate specific DNA binding and an association between Dlx3 and the Msx1 protein in vitro. Immunohistochemical analysis confirmed coexpression of Dlx3 and Msx1 proteins in the differentiated layers of murine epidermal tissues. Transcription factor function requires nuclear localization. In this study, the intracellular localization of the green fluorescent protein fused to Dlx3 was examined in keratinocytes induced to differentiate by calcium and is shown to localize to the nucleus. A bipartite nuclear localization signal (NLS) was identified by mutational analysis and shown to be sufficient for nuclear localization. This was demonstrated by insertion of the Dlx3 bipartite NLS sequence into a cytoplasmic fusion protein, GFP-keratin 14, which functionally redirected GFP-keratin 14 expression to the nucleus. Further analysis of Dlx3 NLS mutants revealed that the Dlx3 NLS sequences are required for specific DNA binding, transactivation potential and interactions with the Msx1 protein.
Figures
Fig. 1
Transactivation potential of Dlx3 and mutated Dlx3 proteins determined by a yeast one-hybrid assay. (A) The GAL4 BD-Dlx3 and GAL4 BD-deletion Dlx3 fusion proteins. The gray boxes represent the GAL4 BD, the white boxes indicate the homeodomain region and the lines represent Dlx3 or truncations of the Dlx3 protein, which correspond to the amino acid numbers indicate on the right. (B) Mean results of three one-hybrid experiments. Each culture was tested in duplicate in the liquid culture β-galactosidase assay. The pattern of transactivation of mutant Dlx3 proteins compared to wild-type Dlx3 was reproducible. β-galactosidase units are an indirect measure of transactivation potential. β-galactosidase units were determined as described in Materials and Methods. Mean β-galactosidase units ± s.d. are recorded on the _x_-axis. GAL4 BD-Dlx3 fusion proteins are recorded on the _y_-axis. GAL4 BD alone is included to indicate the level of background transactivation in the assay.
Fig. 2
Immunoblot of GST-Msx1:Dlx3 pull-down assay. Glutathione-Sepharose 4B resin-bound GST and GST-Msx1 proteins were incubated with Dlx3 proteins. The Msx1:Dlx3 protein complexes were recovered by centrifugation. Immunoblot analysis was performed using IgG anti-Dlx3 antibody at 1:400 dilution to detect Dlx3 proteins in the pull-down assay. The bait proteins, GST-Msx1 and GST, are indicated at the bottom of the figure. Input hexahistidine-Dlx3, Dlx3 1–252 aa and Dlx3 mutated NLS sequences [1+2] proteins combined with the bait proteins in the pull-down reaction are indicated below each lane as Dlx3, 1–252 and [1+2], respectively. Relative molecular mass is indicated in kDa at the left side of the figure.
Fig. 3
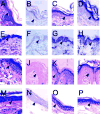
Immunohistochemical analysis of murine neonatal stratified epithelium and adult murine stratified epithelial tissue.(A–D) Murine neonatal epithelium. (E–H) Adult murine trunk epithelial tissue. (I–L) Adult murine whisker pad epithelium. (M–P) Adult murine ear epithelial tissue. (A,E,I,M) Sections were stained with Hematoxylin and Eosin. (B,F,J,N) Immunohistochemical analysis of sections using pre-immune rabbit serum as a negative control for non-specific antibody binding. (C,G,K,O) Immunohistochemical analysis to detect Dlx3 protein. (D,H,L,P) Immunohistochemical analysis to detect Msx1 protein. The basal cell layer is indicated by arrowheads. Original magnification 400×.
Fig. 4
Transient expression of GFP fusion proteins in primary murine keratinocytes induced to differentiate by Ca2+. GFP, GFP-Dlx3 and GFP-Dlx3 mutant fusion proteins were expressed in primary murine keratinocytes growth in 1.4 mM Ca2+ and visualized by direct fluorescence microscopy. (A–C) The same image of a GFP-expressing cell, using (A) an FITC filter, (B) a DAPI filter; (C) a double image of FITC plus DAPI. (D,E) The same image of a GFP-Dlx3 expressing cell, using an FITC filter (D) or a DAPI filter (E); (F) a double image of FITC plus DAPI. The DAPI counterstain indicates the location of the nucleus within the cell. (G–I) Images generated using the FITC filter. (G) A cell expressing GFP-Dlx3 1-172 aa. (H) A cell expressing GFP-Dlx3 1–144 aa. (I) A cell expressing GFP-Dlx3 with the first basic region of the NLS bipartite sequence mutated to alanine and leucine residues ([1]> AAVLL). Original magnification 800×.
Fig. 5
Schematic representation of mutated Dlx3 proteins that were fused to GFP and expressed by transient transfection in primary murine keratinocytes. (A) A schematic representation of full-length Dlx3, 1–287 aa, with the homeodomain sequences depicted by cross-hatched box (130–189aa). The putative bipartite NLS sequence (124–150 aa) is spelled out with the bipartite sequence [1] and sequence [2] enclosed in circles. The amino acids that were substituted with neutral alanine or leucine residues are indicated in bold. (B) The left side of the figure graphically illustrates the 3′ and 5′ Dlx3 truncation mutations, which were cloned and expressed as GFP-mutant Dlx3 fusion proteins. The gray shaded area indicates the location of the homeodomain. The vertical lines and L and A letters indicate the location and substitution residues for the NLS bipartite basic residues of sequence [1] or sequence [2]. The right side of the figure lists the aa of Dlx3 present in each GFP-mutated Dlx3 protein and indicates the location of intracellular expression as determined by direct fluorescence microscopy. +, nuclear fluorescence localization; −, a fluorescence pattern throughout the cell, similar to GFP alone (see Fig. 4). The substitution mutations are designated by the NLS bipartite sequences [1], [2] or [1+2]. The basic residue(s) within that sequence that were substituted are indicated followed by the neutral residues to which the sequences were mutated (i.e. [1]RK>LL). All the basic residues have been substituted in those cases in which the basic residues of sequence [1] or [2] are not listed (i.e. [1]>AAVLL).
Fig. 6
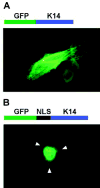
Fluorescence microscopy demonstrating that the Dlx3 NLS is sufficient for nuclear localization. (A) Above the photomicrograph is a schematic representation of the GFP-K14 fusion protein. Below is a direct fluorescence micrograph of a primary murine keratinocyte transiently transfected with pEGFP-C1-K14 and grown for 24 hours in medium containing 0.12 mM Ca2+. (B) Above the photomicrograph is a schematic representation of GFP fused in-framed with the Dlx3 NLS (124–150 aa) fused in-frame with K14. Below is a direct fluorescence micrograph of a primary murine keratinocyte transiently transfected with pEGFP-C1-NLS-K14 and grown for 24 hours in medium containing 0.12 mM Ca2+. Arrowheads indicate the location of the cell membrane.
Fig. 7
Dlx3 NLS sequences and Dlx3 protein function. (A,B) Transactivation potential of Dlx3 1–252 and Dlx3 1–252 NLS mutated proteins determined by a yeast one-hybrid assay. (A) The GAL4 BD-Dlx3 1–252 and GAL4 BD-Dlx3 1–252 NLS mutated fusion proteins. The gray boxes represent the GAL4 BD, the black box indicates the homeodomain region and the lines represent the Dlx3 or mutated Dlx3 sequences that correspond to the amino acid numbers indicate on the left side of B. (B) The averaged results of three one-hybrid experiments. Each culture was tested in duplicate in the liquid culture β-galactosidase assay. The pattern of transactivation of Dlx3 NLS mutant proteins compared to wild-type Dlx3 was reproducible. β-galactosidase units are an indirect measure of transactivation potential, and were determined as described in Materials and Methods. Mean β-galactosidase units ± s.d. are recorded on the _x_-axis. GAL4 BD-Dlx3 fusion proteins are recorded on the _y_-axis. GAL4 BD alone is included to indicate the level of background transactivation in the assay. (C) Gel mobility-shift assays to assess Dlx3 1–252 aa protein and Dlx3 1–252 NLS mutant protein binding to Dlx3 consensus binding site ds oligonucleotide probe DNA. The first lane contains free probe. Lanes 2–5 show gel mobility shifts produced by proteins Dlx3 1–252 aa (Dlx 1–252), Dlx3 1–252 with mutated basic residues in the first NLS bipartite sequence [1], Dlx3 1–252 with mutated basic residues in the second NLS bipartite sequence [2] and Dlx3 1–252 with mutated basic residues in the both NLS bipartite sequences [1+2]. (D) Gel mobility-shift assays to assess Dlx3 and Dlx3 1–252 aa protein binding to Dlx3 consensus binding site ds oligonucleotide probe DNA. The first lane of each photomicrograph contains free probe, followed by a non-competed lane (−). Lanes 3, 4 and 5 contain competition mobility-shift assays with 100-fold excess cold competitor Dlx3 consensus binding site (Dlx), mutated Dlx3 binding site (Mut), and Msx1 consensus binding site (Msx) ds oligonucleotides, respectively. (E) Anti-T7 immunoblot of recombinant hexahistidine-Dlx3 1–252 and Dlx3 1–252 NLS mutant proteins that contain the T7-epitope tag. Lane 1, full-length Dlx3 protein; lane 2, Dlx3 1–252 protein, (Dlx3 1–252); lane 3, Dlx3 1–252 with mutated basic residues in the first NLS bipartite sequence [1]; lane 4, Dlx3 1–252 with mutated basic residues in the second NLS bipartite sequence [2]; lane 5, Dlx3 1–252 with mutated basic residues in the both NLS bipartite sequences [1+2]. Relative molecular mass is indicated in kDa at the left side of E.
Similar articles
- Phosphorylation of murine homeodomain protein Dlx3 by protein kinase C.
Park GT, Denning MF, Morasso MI. Park GT, et al. FEBS Lett. 2001 May 4;496(1):60-5. doi: 10.1016/s0014-5793(01)02398-5. FEBS Lett. 2001. PMID: 11343707 Free PMC article. - Bone morphogenetic protein-2 (BMP-2) transactivates Dlx3 through Smad1 and Smad4: alternative mode for Dlx3 induction in mouse keratinocytes.
Park GT, Morasso MI. Park GT, et al. Nucleic Acids Res. 2002 Jan 15;30(2):515-22. doi: 10.1093/nar/30.2.515. Nucleic Acids Res. 2002. PMID: 11788714 Free PMC article. - Regulation of the Dlx3 homeobox gene upon differentiation of mouse keratinocytes.
Park GT, Morasso MI. Park GT, et al. J Biol Chem. 1999 Sep 10;274(37):26599-608. doi: 10.1074/jbc.274.37.26599. J Biol Chem. 1999. PMID: 10473625 Free PMC article. - Identification of nuclear localization signals in the human homeoprotein MSX1.
Shibata A, Machida J, Yamaguchi S, Kimura M, Tatematsu T, Miyachi H, Nakayama A, Shimozato K, Tokita Y. Shibata A, et al. Biochem Cell Biol. 2018 Aug;96(4):483-489. doi: 10.1139/bcb-2017-0263. Epub 2017 Nov 20. Biochem Cell Biol. 2018. PMID: 29156143 - An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity.
Xiao Z, Latek R, Lodish HF. Xiao Z, et al. Oncogene. 2003 Feb 20;22(7):1057-69. doi: 10.1038/sj.onc.1206212. Oncogene. 2003. PMID: 12592392
Cited by
- Expression and function of Dlx genes in the osteoblast lineage.
Li H, Marijanovic I, Kronenberg MS, Erceg I, Stover ML, Velonis D, Mina M, Heinrich JG, Harris SE, Upholt WB, Kalajzic I, Lichtler AC. Li H, et al. Dev Biol. 2008 Apr 15;316(2):458-70. doi: 10.1016/j.ydbio.2008.01.001. Epub 2008 Jan 16. Dev Biol. 2008. PMID: 18280462 Free PMC article. - Dlx3 is a crucial regulator of hair follicle differentiation and cycling.
Hwang J, Mehrani T, Millar SE, Morasso MI. Hwang J, et al. Development. 2008 Sep;135(18):3149-59. doi: 10.1242/dev.022202. Epub 2008 Aug 6. Development. 2008. PMID: 18684741 Free PMC article. - SUMOylation of DLX3 by SUMO1 promotes its transcriptional activity.
Duverger O, Chen SX, Lee D, Li T, Chock PB, Morasso MI. Duverger O, et al. J Cell Biochem. 2011 Feb;112(2):445-52. doi: 10.1002/jcb.22891. J Cell Biochem. 2011. PMID: 21268066 Free PMC article. - Salt Dependence of DNA Binding Activity of Human Transcription Factor Dlx3.
Jin HS, Son J, Seo YJ, Choi SR, Ahn HB, Go Y, Lim J, Oh KI, Ryu KS, Lee JH. Jin HS, et al. Int J Mol Sci. 2022 Aug 22;23(16):9497. doi: 10.3390/ijms23169497. Int J Mol Sci. 2022. PMID: 36012753 Free PMC article.
References
- Anderson S, Eisenstat D, Shi L, Rubenstein J. Interneuron migration from basal forebrain to neocortex: Dependence on Dlx gene. Science. 1997;278:474–476. - PubMed
- Bendall AJ, Abate-Shen C. Roles for Msx and Dlx homeoproteins in vertebrate development. Gene. 2000;247:17–31. - PubMed
- Billeter M, Qian Y, Otting G, Muller M, Gehring W, Wuthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993;234:1084–1097. - PubMed
- Carriere C, Plaza S, Caboche J, Dozier C, Bailly M, Martin P, Saule S. Nuclear localization signals, DNA binding, and transactivation properties of Quail Pax-6 (Pax-QNR) Isoforms. Cell Growth Diff. 1995;6:1531–1540. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources