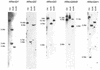Molecular characterisation of RecQ homologues in Arabidopsis thaliana - PubMed (original) (raw)
Molecular characterisation of RecQ homologues in Arabidopsis thaliana
F Hartung et al. Nucleic Acids Res. 2000.
Abstract
Members of the RecQ family of DNA helicases are involved in processes linked to DNA replication, DNA recombination and gene silencing. RecQ homologues of various animals have been described recently. Here, for the first time for plants, we characterised cDNAs of all in all six different RecQ-like proteins that are expressed to different extents in Arabidopsis thaliana. Surprisingly, three of these proteins are small in size [AtRecQl1, AtRecQl2, AtRecQl3-606, 705 and 713 amino acids (aa), respectively], whereas the two bigger proteins result from a duplication event during plant evolution [AtRecQl4A and AtRecQl4B-1150 and 1182 aa, respectively]. Another homologue (AtRecQsim, 858 aa) most probably arose by insertion of an unrelated sequence within its helicase domain. The presence of these homologues demonstrates the conservation of RecQ family functions in higher eukaryotes. We also detected a small gene (AtWRNexo) encoding 285 aa which, being devoid of any RecQ-like helicase domain, reveals a striking homology to the exonuclease domain of human Werner protein, a prominent RecQ helicase of larger size. By means of the two-hybrid assay we were able to detect an interaction between AtWRNexo and AtRecQl2, indicating that activities that reside in a single protein chain in mammals might in plants be complemented in trans.
Figures
Figure 1
Schematic genomic structure of six RecQ-like homologues of A.thaliana. Introns are shown as white boxes within the grey shaded sequence. Introns with a conserved position throughout the six genes are shown as black boxes and are numbered above. The letters below the schematic sequence structure denote the most conserved amino acids of helicase domains II and V (see also Fig. 3).
Figure 2
Schematic structure of RecQ-like proteins from different organisms. The deduced proteins were aligned according to the helicase domains (shown as filled boxes). Stretches of acidic or basic amino acids are represented as light grey or dark grey boxes, respectively. The exonuclease domain of the human WRN protein is indicated. The size of the predicted proteins is given on the right.
Figure 3
Multiple alignment of the seven highly conserved helicase domains of RecQ-like proteins from A.thaliana and various other organisms. The multiple alignment was done with CLUSTALW (version 1.7; www.ibc.wustl.edu ). Gaps are represented by dashes, amino acids conserved in at least eight out of the ten sequences are grey shaded and the consensus amino acid is given in bold below the sequences. The helicase domains are numbered above the sequences. Amino acids considered as conserved in a given position are: (D,E); (K,R); (I,L and V). The organisms included in the alignment are, from top to bottom: E.coli; A.thaliana; N.crassa; H.sapiens; M.musculus and S.cerevisiae.
Figure 4
Southern analysis of the A.thaliana members of the RecQ family. The indicated fragment sizes are calculated from the sequence information of the corresponding genomic BAC clones. All genes show a banding pattern that is in accordance with a presumed single copy status in the Arabidopsis genome.
Figure 5
The small WRNexo gene from A.thaliana. (A) Schematic intron/exon structure of the gene. The five introns are represented as white boxes in the grey shaded sequence. (B) Alignment of AtWRNexo with the N-terminus of the WRN protein from mouse and the X.laevis FFA protein. Amino acids conserved in all three positions are given as letters below the sequence. Highly conserved amino acid stretches are represented as grey shaded boxes. Amino acids considered as conserved in a given position are: (D,E); (K,R); (I,L and V).
Figure 6
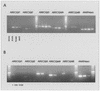
Two-hybrid analysis to detect interactions of AtWRNexo with AtRecQl1 to 3. AtWRNexo and AtRecQl2 showed a strong interaction after 4 h X-gal staining (left). The panels on the right with AtRecQl1 or 3 and AtWRNexo were X-gal stained for 12 h without resulting in a positive signal. As a control the SNF1 + 4 interaction is given in the upper part of the figure.
Figure 7
Comparative RT–PCR of AtRecQl1 to 4B and AtWRNexo using mRNA from different tissues (A) and a quantification of expression in flowers (B). (A) Each cDNA was amplified with 38 PCR cycles starting with reverse transcription of 10 ng mRNA from Arabidopsis rosette leaves, shoots, flowers or seedlings. (B) Each cDNA was amplified with 35 PCR cycles starting with reverse transcription of 10 ng mRNA from Arabidopsis flowers or a 1:10 or 1:100 dilution of this RT, respectively. (A and B) One-fifth of the PCR was subjected to gel electrophoresis and EtBr staining.
Similar articles
- The RecQ gene family in plants.
Hartung F, Puchta H. Hartung F, et al. J Plant Physiol. 2006 Feb;163(3):287-96. doi: 10.1016/j.jplph.2005.10.013. Epub 2005 Dec 20. J Plant Physiol. 2006. PMID: 16371241 Review. - The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Liu Z, Macias MJ, Bottomley MJ, Stier G, Linge JP, Nilges M, Bork P, Sattler M. Liu Z, et al. Structure. 1999 Dec 15;7(12):1557-66. doi: 10.1016/s0969-2126(00)88346-x. Structure. 1999. PMID: 10647186 - Arabidopsis RecQsim, a plant-specific member of the RecQ helicase family, can suppress the MMS hypersensitivity of the yeast sgs1 mutant.
Bagherieh-Najjar MB, de Vries OM, Kroon JT, Wright EL, Elborough KM, Hille J, Dijkwel PP. Bagherieh-Najjar MB, et al. Plant Mol Biol. 2003 May;52(2):273-84. doi: 10.1023/a:1023968429220. Plant Mol Biol. 2003. PMID: 12856935 - Biochemical characterization of an exonuclease from Arabidopsis thaliana reveals similarities to the DNA exonuclease of the human Werner syndrome protein.
Plchova H, Hartung F, Puchta H. Plchova H, et al. J Biol Chem. 2003 Nov 7;278(45):44128-38. doi: 10.1074/jbc.M303891200. Epub 2003 Aug 22. J Biol Chem. 2003. PMID: 12937173
Cited by
- A homolog of ScRAD5 is involved in DNA repair and homologous recombination in Arabidopsis.
Chen IP, Mannuss A, Orel N, Heitzeberg F, Puchta H. Chen IP, et al. Plant Physiol. 2008 Apr;146(4):1786-96. doi: 10.1104/pp.108.116806. Epub 2008 Feb 29. Plant Physiol. 2008. PMID: 18310306 Free PMC article. - Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana.
Hartung F, Suer S, Puchta H. Hartung F, et al. Proc Natl Acad Sci U S A. 2007 Nov 20;104(47):18836-41. doi: 10.1073/pnas.0705998104. Epub 2007 Nov 13. Proc Natl Acad Sci U S A. 2007. PMID: 18000056 Free PMC article. - RecQ helicases: suppressors of tumorigenesis and premature aging.
Bachrati CZ, Hickson ID. Bachrati CZ, et al. Biochem J. 2003 Sep 15;374(Pt 3):577-606. doi: 10.1042/BJ20030491. Biochem J. 2003. PMID: 12803543 Free PMC article. Review. - Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A.
Cui S, Arosio D, Doherty KM, Brosh RM Jr, Falaschi A, Vindigni A. Cui S, et al. Nucleic Acids Res. 2004 Apr 19;32(7):2158-70. doi: 10.1093/nar/gkh540. Print 2004. Nucleic Acids Res. 2004. PMID: 15096578 Free PMC article. - A fluorescence-based exonuclease assay to characterize DmWRNexo, orthologue of human progeroid WRN exonuclease, and its application to other nucleases.
Mason PA, Boubriak I, Cox LS. Mason PA, et al. J Vis Exp. 2013 Dec 23;(82):e50722. doi: 10.3791/50722. J Vis Exp. 2013. PMID: 24378758 Free PMC article.
References
- Karow J.K., Wu,L. and Hickson,I.D. (2000) Curr. Opin. Genet. Dev., 10, 32–38. - PubMed
- Shen J.C. and Loeb,L.A. (2000) Trends Genet., 16, 213–220. - PubMed
- Chakraverty R.K. and Hickson,I.D. (1999) Bioessays, 21, 286–294. - PubMed
- Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) Cell, 83, 655–666. - PubMed
- Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S., Martin,G.M., Mulligan,J. and Schellenberg,G.D. (1996) Science, 272, 258–262. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases