Factors affecting nuclear export of the 60S ribosomal subunit in vivo - PubMed (original) (raw)
Factors affecting nuclear export of the 60S ribosomal subunit in vivo
T Stage-Zimmermann et al. Mol Biol Cell. 2000 Nov.
Free PMC article
Abstract
In Saccharomyces cerevisiae, the 60S ribosomal subunit assembles in the nucleolus and then is exported to the cytoplasm, where it joins the 40S subunit for translation. Export of the 60S subunit from the nucleus is known to be an energy-dependent and factor-mediated process, but very little is known about the specifics of its transport. To begin to address this problem, an assay was developed to follow the localization of the 60S ribosomal subunit in S. cerevisiae. Ribosomal protein L11b (Rpl11b), one of the approximately 45 ribosomal proteins of the 60S subunit, was tagged at its carboxyl terminus with the green fluorescent protein (GFP) to enable visualization of the 60S subunit in living cells. A panel of mutant yeast strains was screened for their accumulation of Rpl11b-GFP in the nucleus as an indicator of their involvement in ribosome synthesis and/or transport. This panel included conditional alleles of several rRNA-processing factors, nucleoporins, general transport factors, and karyopherins. As predicted, conditional alleles of rRNA-processing factors that affect 60S ribosomal subunit assembly accumulated Rpl11b-GFP in the nucleus. In addition, several of the nucleoporin mutants as well as a few of the karyopherin and transport factor mutants also mislocalized Rpl11b-GFP. In particular, deletion of the previously uncharacterized karyopherin KAP120 caused accumulation of Rpl11b-GFP in the nucleus, whereas ribosomal protein import was not impaired. Together, these data further define the requirements for ribosomal subunit export and suggest a biological function for KAP120.
Figures
Figure 1
The RPL11B–GFP gene fusion is functional in yeast. (A) Four spores of a representative tetrad from the cross between PSY2083 (rpl11a::HIS3) covered by YCp50L11A URA3 CEN and PSY2084 (rpl11b::HIS3) streaked on synthetic complete medium containing FOA (left panel). Spores a and d are HIS+ and FOAS and spores b and c are his− and FOAR. Spores a–d were transformed with pPS2167 (RPL11B–GFP LEU2 CEN) and streaked on synthetic complete medium containing FOA (right panel). (B) Rpl11b–GFP expressed from a CEN plasmid in wild-type yeast was visualized by fluorescence microscopy of living cells (left panel). Arrows point to a vacuole (v) and the nucleus (n). Nomarski, phase-contrast image of cells; Rpl11b–GFP, fluorescence signal. Yeast lysate prepared from wild-type yeast cells expressing Rpl11b–GFP was probed with anti-GFP to detect Rpl11b–GFP (right panel). Five micrograms of total protein of lysate was loaded. Migration of a 10-kDa ladder of molecular mass markers is shown.
Figure 2
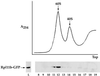
Rpl11b–GFP is incorporated in the 60S ribosomal subunit. Yeast lysate from wild-type cells expressing Rpl11b–GFP from a CEN plasmid was separated on a 7–30% sucrose gradient. The sucrose gradient profile was recorded by absorbance at 254 nm (upper panel). Fractions 8–19 collected from the sucrose gradient and lysate (5 μg of total protein) were probed with anti-GFP to detect Rpl11b–GFP (bottom panel).
Figure 3
Rpl11b–GFP localization assay. Wild-type (PSY580) or mtr4-1 cells expressing Rpl11b–GFP from a CEN plasmid were grown to early log phase, shifted to 37°C for 1 h, and then shifted back to 25°C for up to 1 h. Yeast cells were fixed and incubated with DAPI to stain the nuclei. Rpl11b–GFP was visualized by fluorescence microscopy. Panels a, b, e, and f show cells shifted to 37°C for 1 h, and panels c, d, g, and h show cells shifted back to 25°C for 30 min.
Figure 4
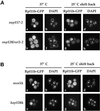
Rpl11b–GFP localization in nucleoporin and karyopherin mutants. Yeast strains nup157-2,rat2-2, msn5Δ, and_kap120Δ_ expressing Rpl11b–GFP from a CEN plasmid were grown to early log phase, shifted to 37°C for 1 h, and then shifted back to 25°C for up to 1 h. Cells were fixed, and their nuclei were stained with DAPI. (A) Fluorescence microscopy images of_nup157-2_ cells shifted to 37°C (a and b),nup157-2 cells shifted back to 25°C for 30 min (c and d), rat2-2 cells shifted to 37°C (e and f), and_rat2-2_ cells 30 min after shifting back to 25°C (g and h). (B) Fluorescence microscopy images of msn5Δ cells shifted to 37°C (a and b), msn5Δ cells shifted back to 25°C for 30 min (c and d), kap120Δ cells shifted to 37°C (e and f), and kap120Δ cells 30 min after shifting back to 25°C (g and h).
Figure 5
Processing of rRNA in KAP120 and_kap120Δ_ cells. KAP120 and_kap120Δ_ cells were grown at 25°C to a density of 1 × 107 cells/ml, pulse labeled for 3 min with [3H-methyl]methionine, and chased with an excess of unlabeled methionine. Chase time points of 1, 3, and 10 min for each yeast strain are shown. The 35S rRNA precursor and processing intermediates are labeled.
Figure 6
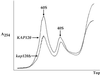
Deletion of KAP120 causes a large reduction in 60S ribosomal subunit levels. Yeast lysates prepared from_KAP120_ and kap120Δ cells expressing Rpl11b–GFP from a CEN plasmid were centrifuged through 7–30% sucrose gradients, and gradient profiles were collected by absorbance at 254 nm. An overlay of the gradient profiles for KAP120 and_kap120Δ_ cells is shown to emphasize the difference in peak heights for the 60S and 40S ribosomal subunits. Equal amounts of total RNA (200 μg) were loaded for each gradient.
Similar articles
- A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants.
Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. Hurt E, et al. J Cell Biol. 1999 Feb 8;144(3):389-401. doi: 10.1083/jcb.144.3.389. J Cell Biol. 1999. PMID: 9971735 Free PMC article. - Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p.
Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Gadal O, et al. Mol Cell Biol. 2001 May;21(10):3405-15. doi: 10.1128/MCB.21.10.3405-3415.2001. Mol Cell Biol. 2001. PMID: 11313466 Free PMC article. - Intersection of the Kap123p-mediated nuclear import and ribosome export pathways.
Sydorskyy Y, Dilworth DJ, Yi EC, Goodlett DR, Wozniak RW, Aitchison JD. Sydorskyy Y, et al. Mol Cell Biol. 2003 Mar;23(6):2042-54. doi: 10.1128/MCB.23.6.2042-2054.2003. Mol Cell Biol. 2003. PMID: 12612077 Free PMC article. - Nuclear export of ribosomal subunits.
Johnson AW, Lund E, Dahlberg J. Johnson AW, et al. Trends Biochem Sci. 2002 Nov;27(11):580-5. doi: 10.1016/s0968-0004(02)02208-9. Trends Biochem Sci. 2002. PMID: 12417134 Review. - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
Cited by
- Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae.
Seiser RM, Sundberg AE, Wollam BJ, Zobel-Thropp P, Baldwin K, Spector MD, Lycan DE. Seiser RM, et al. Genetics. 2006 Oct;174(2):679-91. doi: 10.1534/genetics.106.062117. Epub 2006 Aug 3. Genetics. 2006. PMID: 16888326 Free PMC article. - A novel conserved RNA-binding domain protein, RBD-1, is essential for ribosome biogenesis.
Björk P, Baurén G, Jin S, Tong YG, Bürglin TR, Hellman U, Wieslander L. Björk P, et al. Mol Biol Cell. 2002 Oct;13(10):3683-95. doi: 10.1091/mbc.e02-03-0138. Mol Biol Cell. 2002. PMID: 12388766 Free PMC article. - Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export.
Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Brune C, et al. RNA. 2005 Apr;11(4):517-31. doi: 10.1261/rna.7291205. RNA. 2005. PMID: 15769879 Free PMC article. - Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis.
Oeffinger M, Fatica A, Rout MP, Tollervey D. Oeffinger M, et al. Nucleic Acids Res. 2007;35(4):1354-66. doi: 10.1093/nar/gkl824. Epub 2007 Feb 1. Nucleic Acids Res. 2007. PMID: 17272295 Free PMC article. - Retroviruses and yeast retrotransposons use overlapping sets of host genes.
Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, Liou W, Sandmeyer S. Irwin B, et al. Genome Res. 2005 May;15(5):641-54. doi: 10.1101/gr.3739005. Epub 2005 Apr 18. Genome Res. 2005. PMID: 15837808 Free PMC article.
References
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113:1651–1659. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials