Maturation of IncP pilin precursors resembles the catalytic Dyad-like mechanism of leader peptidases - PubMed (original) (raw)
Maturation of IncP pilin precursors resembles the catalytic Dyad-like mechanism of leader peptidases
R Eisenbrandt et al. J Bacteriol. 2000 Dec.
Abstract
The pilus subunit, the pilin, of conjugative IncP pili is encoded by the trbC gene. IncP pilin is composed of 78 amino acids forming a ring structure (R. Eisenbrandt, M. Kalkum, E.-M. Lai, C. I. Kado, and E. Lanka, J. Biol. Chem. 274:22548-22555, 1999). Three enzymes are involved in maturation of the pilin: LepB of Escherichia coli for signal peptide removal and a yet-unidentified protease for removal of 27 C-terminal residues. Both enzymes are chromosome encoded. Finally, the inner membrane-associated IncP TraF replaces a four-amino-acid C-terminal peptide with the truncated N terminus, yielding the cyclic polypeptide. We refer to the latter process as "prepilin cyclization." We have used site-directed mutagenesis of trbC and traF to unravel the pilin maturation process. Each of the mutants was analyzed for its phenotypes of prepilin cyclization, pilus formation, donor-specific phage adsorption, and conjugative DNA transfer abilities. Effective prepilin cyclization was determined by matrix-assisted laser desorption-ionization-mass spectrometry using an optimized sample preparation technique of whole cells and trans-3-indolyl acrylic acid as a matrix. We found that several amino acid exchanges in the TrbC core sequence allow prepilin cyclization but disable the succeeding pilus assembly. We propose a mechanism explaining how the signal peptidase homologue TraF attacks a C-terminal section of the TrbC core sequence via an activated serine residue. Rather than cleaving and releasing hydrolyzed peptides, TraF presumably reacts as a peptidyl transferase, involving the N terminus of TrbC in the aminolysis of a postulated TraF-acetyl-TrbC intermediate. Under formal loss of a C-terminal tetrapeptide, a new peptide bond is formed in a concerted action, connecting serine 37 with glycine 114 of TrbC.
Figures
FIG. 1
Processing scheme of RP4 TrbC. The protein is shown as a box with its sequence in one-letter code inside. The 36-aa signal peptide and the 27-aa C-terminal cleaved peptide are shown as white boxes. The core sequence is shaded yellow with two predicted transmembranal helices (TMH) shown in blue. The 4-aa residues removed by TraF are inverted. Point mutations are annotated below or above the original sequence. Letters below indicate point mutants that are still cyclized by TraF; letters above indicate point mutants that are not processed by TraF. LepB and TraF are the enzymes that cleave TrbC, where “?” is the as-yet-unidentified host-encoded protease.
FIG. 2
Sequence alignment of TraF-like proteins. Amino acid compositions of the different gene products are arranged in families; GenBank accession numbers are given in parentheses. Dark yellow background, bacterial conjugative plasmids' RP4 (L27758) and R751 (M94367) TraF. Yellow background, A. tumefaciens TraF homologues from pTi15955 (P15595), pTiA6 (U43674), pTiC58 (U40389), and putative pTi15955 protein Orf2 (S15913), pTiA6NC VirI (2773263), as well as Rhizobium Ti plasmid homologue pNGR234a TraF (P55417). Light yellow background, leader peptidases LepB of E. coli (K00426), Lep of Salmonella enterica serovar Typhimurium (X54933), Lep of Synechocystis sp. (PCC6803), SipS of B. subtilis (Z11847), SipP of _B. subtilis_plasmid pTA1015 (AAC44415), SpsB of Staphylococcus aureus(U65000), and Lep of Phormidium laminosum (S51921). Pale yellow background, N. meningitidis PilC1 and PilC2 (Y13020 and Y13021, respectively). Identical amino acid residues in at least 11 sequences are shown with a red background. The catalytically active residues shown for E. coli leader peptidase I are marked with a filled rhombus, the respective amino acids in RP4 TraF have been mutated, and further mutation sites in RP4 TraF are marked with an open rhombus, whereas the predicted transmembranal helix (TMH) for RP4/R751 TraF is indicated with a line above the alignment. Gaps introduced to maximize alignment are indicated by dots. To indicate the high conservation of TraF-like proteins in the uppermost two families, identical amino acid residues of these proteins are indicated by a blue background.
FIG. 3
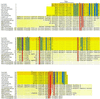
Western blot analysis (for conditions, see Materials and Methods) of E. coli SCS1 cell extracts (2 μl/lane) containing the plasmids indicated. (A) Mutations in_trbC_. Lane a, pRE178 (trbC+) and pJH472 (traF+); lane b, pRE178S37C and pJH472; lane c, pRE178S37P and pJH472; lane d, pRE178F79Δ and pJH472; lane e, pRE178G112D and pJH472; lane f, pRE178R113A and pJH472; lane g, pRE178G114A and pJH472; lane h, pRE178G114S and pJH472; lane i, pRE178G114T and pJH472; lane j, pRE178A115G and pJH472; lane k, pRE178A115S and pJH472; lane l, pRE178E116Q and pJH472; lane m, pRE178I117H and pJH472; lane n, pMS119 (vector) and pJH472. (B) Mutations in traF. Lane a′, pRE178 (trbC+) and pJH472 (traF+); lane b′, pRE178 and pJH472S37A; lane c′, pRE178 and pJH472C59A; lane d′, pRE178 and pJH472C80A; lane e′, pRE178 and pJH472K89Q; lane f′, pRE178 and pJH472K89L; lane g′, pRE178 and pJH472K89R; lane h′, pRE178 and pJH472R90L; lane i′, pRE178 and pJH472P129I; lane j′, pRE178 and pJH472D155I; lane k′, pRE178 and pJH472D155N; lane l′, pRE178 and pJH472R157A; lane m′, pRE178 and pJH472Y158F; lane n′, pMS119EH (vector) and pJH472. Positions of the 145-aa PreProTrbC, N-terminally cleaved ProTrbC, N- and C-terminally processed TrbC∗, and (circular) pilin are indicated on the right side of the figure.
FIG. 4
Western blot analysis (for conditions see Materials and Methods) of E. coli SCS1 cell extracts (2 μl/lane) in the absence (left) or the presence (right) of_traF_. Plasmids used are as follows. Lane a, pMS119EH (vector); lane b, pRE178 (trbC+); lane c, pRE178Δ3 (_trbC_Δ3+); lane d, pRE178Δ3.05 (_trbC_Δ3.05+); lane e, pRE178Δ3.1 (_trbC_Δ3.1+); lane f, pRE178Δ4 (_trbC_Δ4+); lane a′, pJH472 (traF+) and pMS119EH (vector); lane b′, pJH472 (traF+) and pRE178 (trbC+); lane c′, pJH472 (traF+) and pRE178Δ3 (_trbC_Δ3+); lane d′, pJH472 (traF+) and pRE178Δ3.05 (_trbC_Δ3.05+); lane e′, pJH472 (traF+) and pRE178Δ3.1 (_trbC_Δ3.1+); lane f′, pJH472 (traF+) and pRE178Δ4 (_trbC_Δ4+). Standard molecular mass markers (Rainbow labeled markers, low range; Amersham Pharmacia Biotech): Lys, lysozyme (14.3 kDa), and Apr, aprotinin (6.5 kDa). Positions of the 145-aa PreProTrbC, N-terminally cleaved ProTrbC, N- and C-terminally processed TrbC∗, and (circular) pilin (TrbC) are indicated on the right side of the figure.
FIG. 5
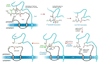
Proposed mechanism for the TraF-catalyzed formation of an internal peptide bond in TrbC. For details, see Discussion.
FIG. 6
Potential common processing mechanism of RP4 TrbC (M93696), Bordetella pertussis PtlA (L10720), Brucella suis VirB2 (AF141604), and Brucella abortus VirB2 (AF226278). The latter two sequences are identical and shown once only. Databank GenBank accession numbers are given in parentheses. Only two short portions of the respective sequences are shown. The first and last amino acid positions in each line are numbered according to the original sequences. Lines correspond to conserved residues in all sequences; colons in only two of the sequences. Residues of a proposed common maturation process are printed in white. Arrows mark sites of RP4 TrbC processing.
Similar articles
- Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits.
Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CI, Lanka E. Eisenbrandt R, et al. J Biol Chem. 1999 Aug 6;274(32):22548-55. doi: 10.1074/jbc.274.32.22548. J Biol Chem. 1999. PMID: 10428832 - A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis.
Haase J, Lanka E. Haase J, et al. J Bacteriol. 1997 Sep;179(18):5728-35. doi: 10.1128/jb.179.18.5728-5735.1997. J Bacteriol. 1997. PMID: 9294428 Free PMC article. - Kinetics and sequence specificity of processing of prepilin by PilD, the type IV leader peptidase of Pseudomonas aeruginosa.
Strom MS, Lory S. Strom MS, et al. J Bacteriol. 1992 Nov;174(22):7345-51. doi: 10.1128/jb.174.22.7345-7351.1992. J Bacteriol. 1992. PMID: 1429457 Free PMC article. - Posttranslational processing of type IV prepilin and homologs by PilD of Pseudomonas aeruginosa.
Strom MS, Nunn DN, Lory S. Strom MS, et al. Methods Enzymol. 1994;235:527-40. doi: 10.1016/0076-6879(94)35168-6. Methods Enzymol. 1994. PMID: 8057924 Review. - Tying rings for sex.
Kalkum M, Eisenbrandt R, Lurz R, Lanka E. Kalkum M, et al. Trends Microbiol. 2002 Aug;10(8):382-7. doi: 10.1016/s0966-842x(02)02399-5. Trends Microbiol. 2002. PMID: 12160637 Review.
Cited by
- Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system.
Kerr JE, Christie PJ. Kerr JE, et al. J Bacteriol. 2010 Oct;192(19):4923-34. doi: 10.1128/JB.00557-10. Epub 2010 Jul 23. J Bacteriol. 2010. PMID: 20656905 Free PMC article. - Biological diversity of prokaryotic type IV secretion systems.
Alvarez-Martinez CE, Christie PJ. Alvarez-Martinez CE, et al. Microbiol Mol Biol Rev. 2009 Dec;73(4):775-808. doi: 10.1128/MMBR.00023-09. Microbiol Mol Biol Rev. 2009. PMID: 19946141 Free PMC article. Review. - Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27.
Lawley TD, Gilmour MW, Gunton JE, Standeven LJ, Taylor DE. Lawley TD, et al. J Bacteriol. 2002 Apr;184(8):2173-80. doi: 10.1128/JB.184.8.2173-2180.2002. J Bacteriol. 2002. PMID: 11914349 Free PMC article. - Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds.
McIntosh JA, Donia MS, Schmidt EW. McIntosh JA, et al. Nat Prod Rep. 2009 Apr;26(4):537-59. doi: 10.1039/b714132g. Nat Prod Rep. 2009. PMID: 19642421 Free PMC article. Review. - The structural biology of type IV secretion systems.
Fronzes R, Christie PJ, Waksman G. Fronzes R, et al. Nat Rev Microbiol. 2009 Oct;7(10):703-14. doi: 10.1038/nrmicro2218. Nat Rev Microbiol. 2009. PMID: 19756009 Free PMC article. Review.
References
- Anthony K G, Sherburne C, Sherburne R, Frost L S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13:939–953. - PubMed
- Bamford D H, Rouhiainen L, Takkinen K, Söderlund H. Comparison of the lipid-containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J Gen Virol. 1981;57:365–373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources